Rearranged Oriloc output example: Previous Index Next
Bacteria; Actinobacteria; Actinobacteridae; Actinomycetales; Corynebacterineae; Corynebacteriaceae; Corynebacterium.
Genome size (bp): 3147090
Number of genes 2950
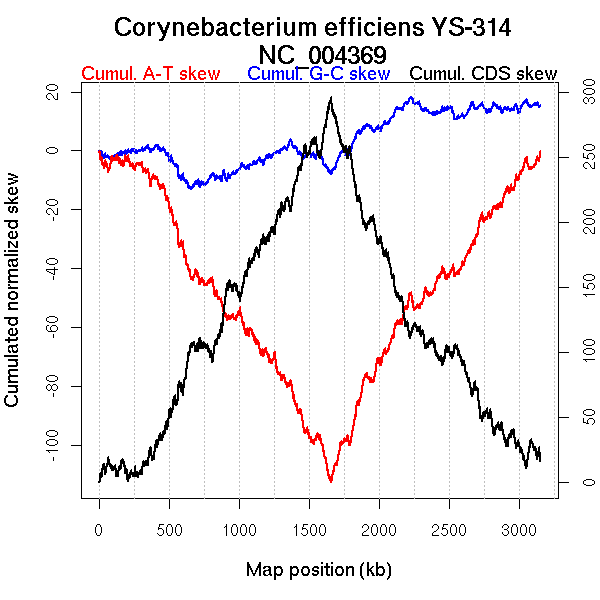
Oriloc predictions: Origin 0 kb Terminus 1653 kb
Worning et al., 2006: Origin 3045 kb Terminus 1654 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 31.811 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.86 kb
Consensus predictions: Origin 0 kb Terminus 1653 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 1047 | NA | 2024 |
| GC-skew reverse | 2254 | NA | 1858 |
| AT-skew forward | 1121 | NA | 2252 |
| AT-skew reverse | 1699 | NA | 533 |
| 2120 | NA | 1637 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 1047 (2024.115 kb) | leading | -0.12 |
| 1048 (2028.468 kb) | 1483 (3146.959 kb) | lagging | -0.165 | |
| GC-skew reverse | 1484 (0 kb) | 2254 (1857.714 kb) | NA | 0.173 |
| 2255 (1859.058 kb) | 2950 (3146.959 kb) | lagging | 0.126 | |
| AT-skew forward | 1 (0 kb) | 1121 (2252.413 kb) | NA | -0.276 |
| 1122 (2253.512 kb) | 1483 (3146.959 kb) | lagging | -0.239 | |
| AT-skew reverse | 1484 (0 kb) | 1699 (532.6115 kb) | leading | 0.231 |
| 1700 (540.696 kb) | 2120 (1637.2635 kb) | leading | 0.239 | |
| 2121 (1642.051 kb) | 2950 (3146.959 kb) | lagging | 0.299 |
More G than C on the leading strand for replication - for forward encoded genes.