Rearranged Oriloc output example: Previous Index Next
Bacteria; Actinobacteria; Actinobacteridae; Actinomycetales; Micrococcineae; Cellulomonadaceae; Tropheryma.
Genome size (bp): 925938
Number of genes 780
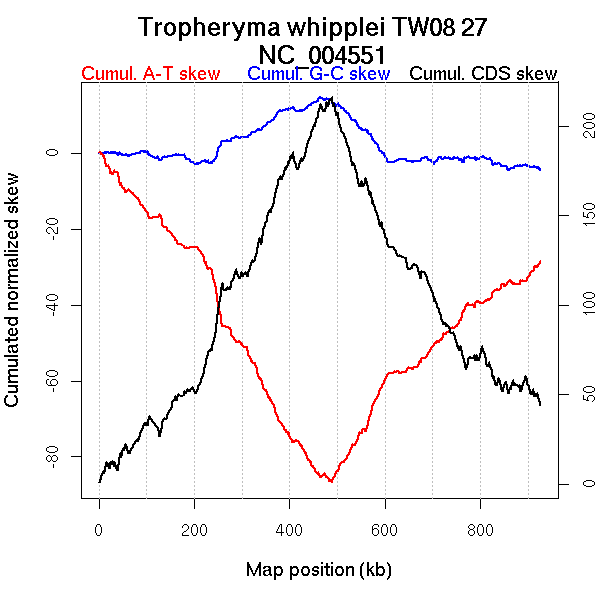
Oriloc predictions: Origin 3 kb Terminus 484 kb
Worning et al., 2006: Origin 478 kb Terminus 4 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 1.479 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.72 kb
Consensus predictions: Origin 3 kb Terminus 484 kb
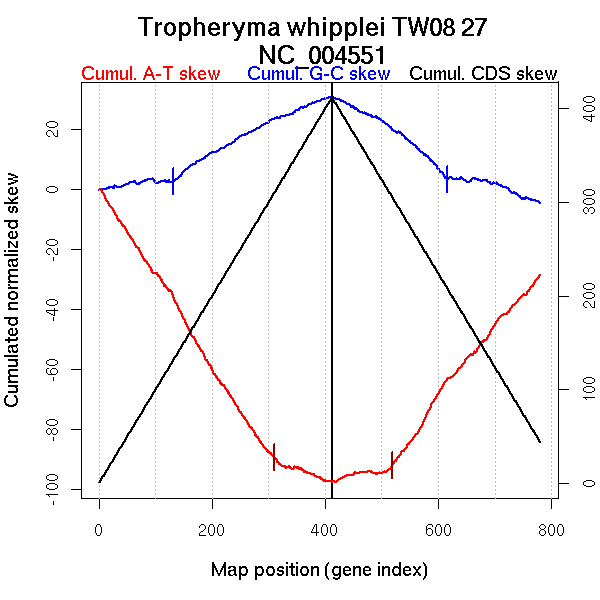
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 131 | 0.00667 | 239 |
| GC-skew reverse | 616 | 0.04 | 620 |
| AT-skew forward | 310 | 0 | 476 |
| AT-skew reverse | 518 | 0 | 491 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 131 (239.1485 kb) | leading | 0.024 |
| 132 (240.175 kb) | 412 (925.735 kb) | NA | 0.098 | |
| GC-skew reverse | 413 (0 kb) | 616 (619.615 kb) | NA | -0.129 |
| 617 (620.812 kb) | 780 (925.735 kb) | lagging | -0.055 | |
| AT-skew forward | 1 (0 kb) | 310 (476.237 kb) | leading | -0.298 |
| 311 (476.6185 kb) | 412 (925.735 kb) | lagging | -0.07 | |
| AT-skew reverse | 413 (0 kb) | 518 (490.7535 kb) | leading | 0.033 |
| 519 (491.3895 kb) | 780 (925.735 kb) | lagging | 0.241 |