Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Alphaproteobacteria; Rickettsiales; Anaplasmataceae; Ehrlichia.
Genome size (bp): 1499920
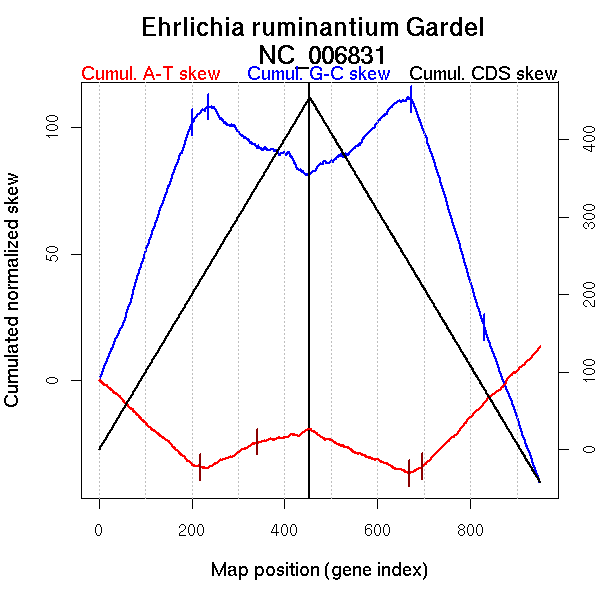
Number of genes 950
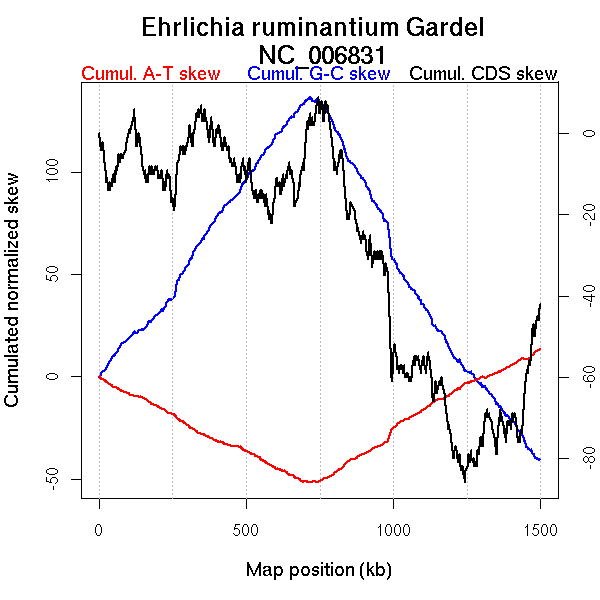
Oriloc predictions: Origin 0 kb Terminus 716 kb
Worning et al., 2006: Origin 1496 kb Terminus 701 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 4.28 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 484.36 kb
Consensus predictions: Origin 0 kb Terminus 716 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 201 | NA | 684 |
| 235 | NA | 754 | |
| GC-skew reverse | 672 | NA | 715 |
| 830 | NA | 1105 | |
| AT-skew forward | 219 | NA | 710 |
| 340 | NA | 1209 | |
| AT-skew reverse | 668 | NA | 693 |
| 697 | NA | 789 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 201 (684.444 kb) | leading | 0.524 |
| 202 (685.5225 kb) | 235 (754.001 kb) | NA | 0.171 | |
| 236 (755.8925 kb) | 454 (1499.902 kb) | lagging | -0.115 | |
| GC-skew reverse | 455 (0 kb) | 672 (714.5835 kb) | leading | 0.151 |
| 673 (718.888 kb) | 830 (1105.1435 kb) | lagging | -0.589 | |
| 831 (1107.057 kb) | 950 (1499.902 kb) | lagging | -0.51 | |
| AT-skew forward | 1 (0 kb) | 219 (709.603 kb) | leading | -0.166 |
| 220 (713.4405 kb) | 340 (1209.259 kb) | lagging | 0.091 | |
| 341 (1217.9525 kb) | 454 (1499.902 kb) | lagging | 0.041 | |
| AT-skew reverse | 455 (0 kb) | 668 (692.678 kb) | leading | -0.078 |
| 669 (694.3815 kb) | 697 (789.1565 kb) | lagging | 0.091 | |
| 698 (790.8825 kb) | 950 (1499.902 kb) | lagging | 0.187 |
More G than C on the leading strand for replication.