Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Cupriavidus.
Genome size (bp): 2726152
Number of genes 2407
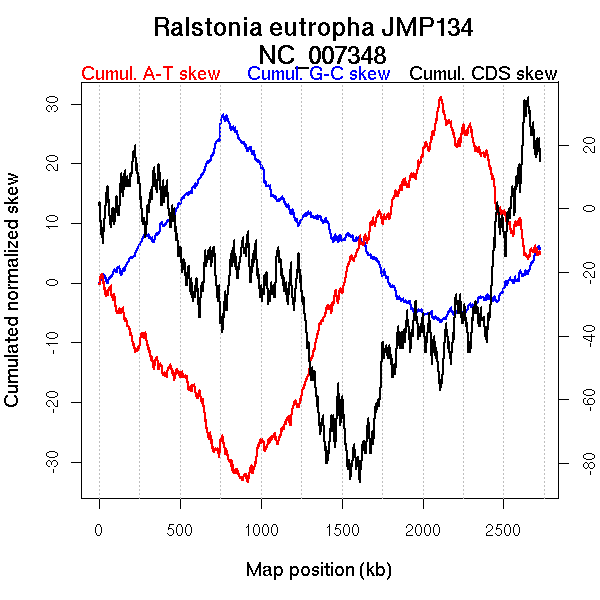
Oriloc predictions: Origin 2107 kb Terminus 788 kb
Worning et al., 2006: Origin 2121 kb Terminus 767 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 262.041 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): NA
Consensus predictions: Origin 2107 kb Terminus 788 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 311 | 0.02 | 766 |
| 853 | 0.02667 | 1961 | |
| GC-skew reverse | 1580 | 0.00667 | 836 |
| 2174 | 0.00667 | 2145 | |
| AT-skew forward | 351 | 0 | 829 |
| AT-skew reverse | 1614 | 0.00333 | 923 |
| 2178 | 0.01667 | 2152 |
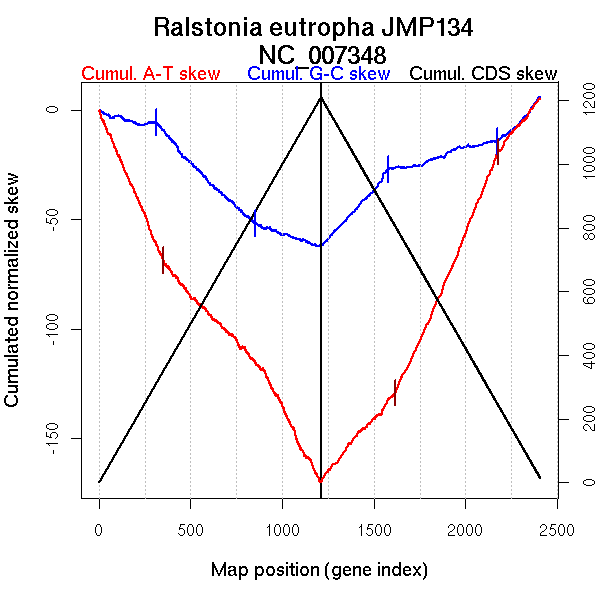
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 311 (766.199 kb) | leading | -0.019 |
| 312 (767.05 kb) | 853 (1961.2345 kb) | lagging | -0.085 | |
| 854 (1966.261 kb) | 1211 (2725.616 kb) | leading | -0.029 | |
| GC-skew reverse | 1212 (0 kb) | 1580 (836.1595 kb) | leading | 0.093 |
| 1581 (837.1095 kb) | 2174 (2145.165 kb) | lagging | 0.025 | |
| 2175 (2147.6375 kb) | 2407 (2725.616 kb) | leading | 0.086 | |
| AT-skew forward | 1 (0 kb) | 351 (828.8445 kb) | leading | -0.194 |
| 352 (829.4665 kb) | 1211 (2725.616 kb) | NA | -0.112 | |
| AT-skew reverse | 1212 (0 kb) | 1614 (922.603 kb) | leading | 0.097 |
| 1615 (922.996 kb) | 2178 (2151.5915 kb) | lagging | 0.197 | |
| 2179 (2154.0095 kb) | 2407 (2725.616 kb) | leading | 0.105 |
More G than C on the leading strand for replication.