Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Pseudoalteromonadaceae; Pseudoalteromonas.
Genome size (bp): 3214944
Number of genes 2939
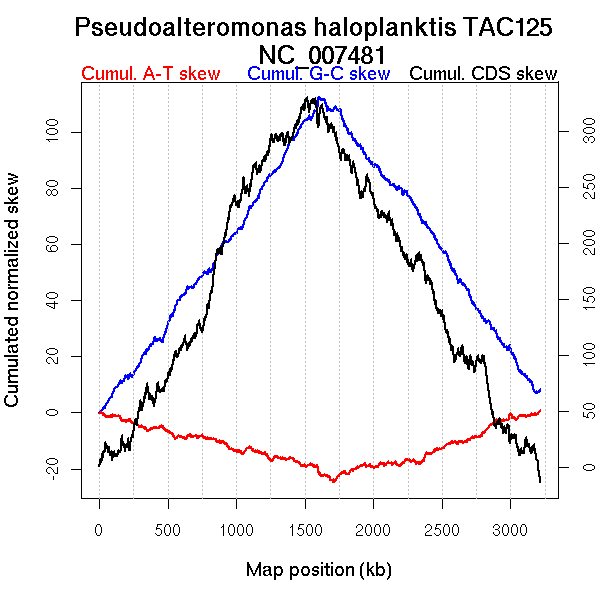
Oriloc predictions: Origin 3179 kb Terminus 1611 kb
Worning et al., 2006: Origin 0 kb Terminus 1623 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 779.835 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): NA
Consensus predictions: Origin 3179 kb Terminus 1611 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 921 | NA | 1631 |
| GC-skew reverse | 2120 | NA | 1723 |
| AT-skew forward | 422 | NA | 777 |
| 496 | NA | 862 | |
| AT-skew reverse | 2029 | NA | 1577 |
| 2102 | NA | 1693 | |
| 2411 | NA | 2262 |
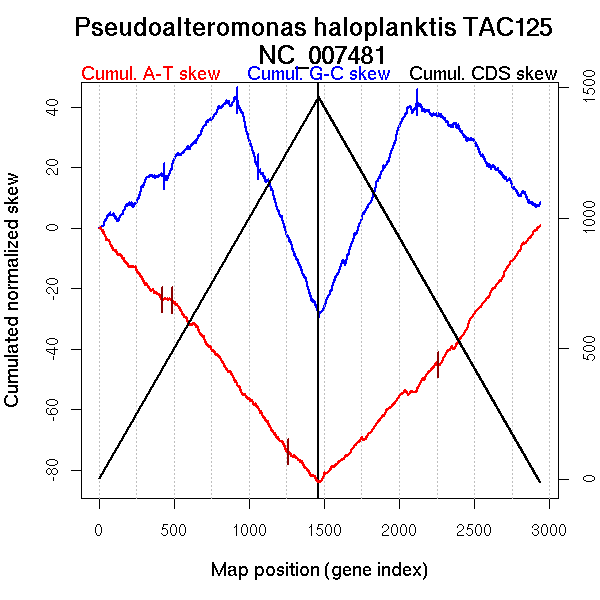
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 921 (1630.574 kb) | leading | 0.049 |
| 922 (1638.0575 kb) | 1463 (3214.586 kb) | lagging | -0.126 | |
| GC-skew reverse | 1464 (0 kb) | 2120 (1723.3805 kb) | leading | 0.114 |
| 2121 (1725.665 kb) | 2939 (3214.586 kb) | lagging | -0.045 | |
| AT-skew forward | 1 (0 kb) | 422 (777.1425 kb) | leading | -0.055 |
| 423 (779.2505 kb) | 496 (861.509 kb) | leading | -0.005 | |
| 497 (864.183 kb) | 1463 (3214.586 kb) | NA | -0.064 | |
| AT-skew reverse | 1464 (0 kb) | 2029 (1576.5905 kb) | leading | 0.052 |
| 2030 (1577.2865 kb) | 2102 (1693.243 kb) | NA | -0.003 | |
| 2103 (1694.248 kb) | 2411 (2261.8155 kb) | lagging | 0.055 | |
| 2412 (2264.6365 kb) | 2939 (3214.586 kb) | lagging | 0.07 |
More G than C on the leading strand for replication.