Rearranged Oriloc output example: Previous Index Next
Bacteria; Firmicutes; Mollicutes; Acholeplasmatales; Acholeplasmataceae; Candidatus Phytoplasma; Candidatus Phytoplasma asteris.
Genome size (bp): 706569
Number of genes 671
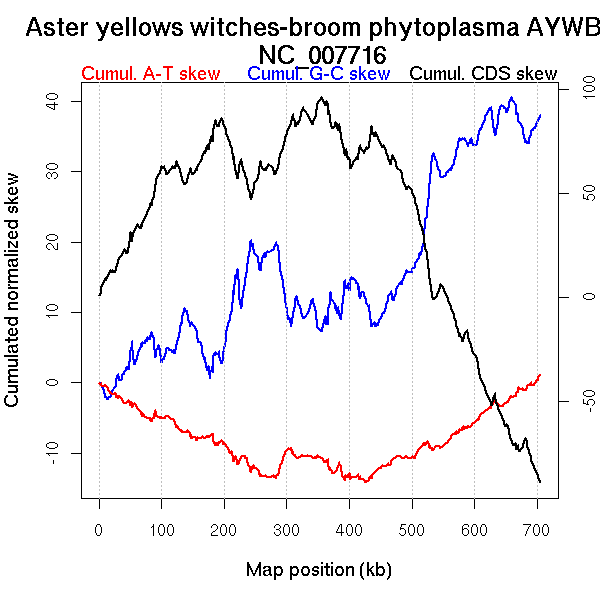
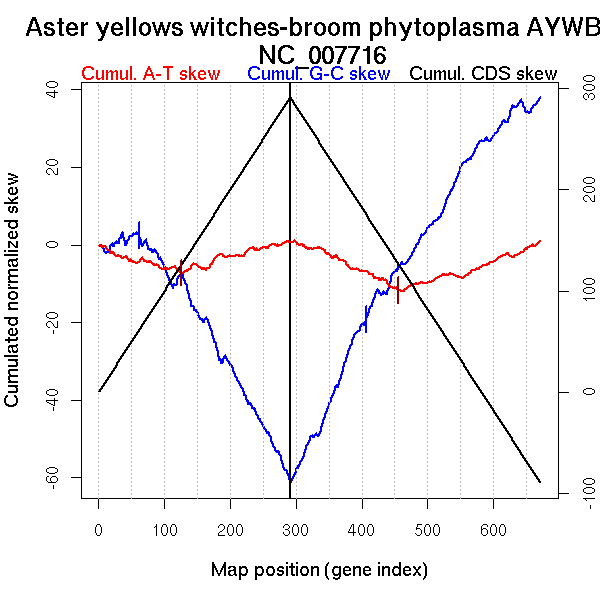
Oriloc predictions: Origin 0 kb Terminus 393 kb
Worning et al., 2006: Origin 705 kb Terminus 446 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 149.369 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.76 kb, 81.68 kb
Consensus predictions: Origin 0 kb Terminus 393 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 61 | 0.02 | 88 |
| GC-skew reverse | 406 | 0 | 337 |
| AT-skew forward | 126 | 0.02667 | 191 |
| AT-skew reverse | 456 | 0 | 402 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 61 (87.868 kb) | leading | 0.076 |
| 62 (88.4665 kb) | 291 (705.696 kb) | NA | -0.276 | |
| GC-skew reverse | 292 (0 kb) | 406 (336.9875 kb) | leading | 0.384 |
| 407 (338.89 kb) | 671 (705.696 kb) | lagging | 0.227 | |
| AT-skew forward | 1 (0 kb) | 126 (191.3645 kb) | leading | -0.048 |
| 127 (192.708 kb) | 291 (705.696 kb) | NA | 0.047 | |
| AT-skew reverse | 292 (0 kb) | 456 (401.617 kb) | leading | -0.078 |
| 457 (407.615 kb) | 671 (705.696 kb) | lagging | 0.056 |
More G than C on the leading strand for replication - for reverse encoded genes.