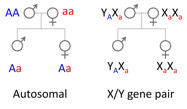
The human XY chromosomes are the best studied sex chromosomes, with plenty of available data. They originated ~180 MY ago in the ancestor of marsupials and placentals with the emergence of Sry, the master male-determining gene. They underwent a series of recombination suppression events, the non-recombining Y lost most of its genes and the X evolved a rescue mechanism compensating for Y gene loss. We have been working on one unclear aspect of this scenario: how and why X-Y recombination is suppressed.
One hypothesis posits that sexually-antagonistic mutations drives recombination suppression. In primates, sexual dimorphism is strong is monkeys and apes and very weak in lemurs and lorises. We know of lot about sex chromosome evolution in the former, but almost nothing in the latter. We have set up a study of the sex chromosomes in lemurs and lorises top test the above-mentioned hypothesis.