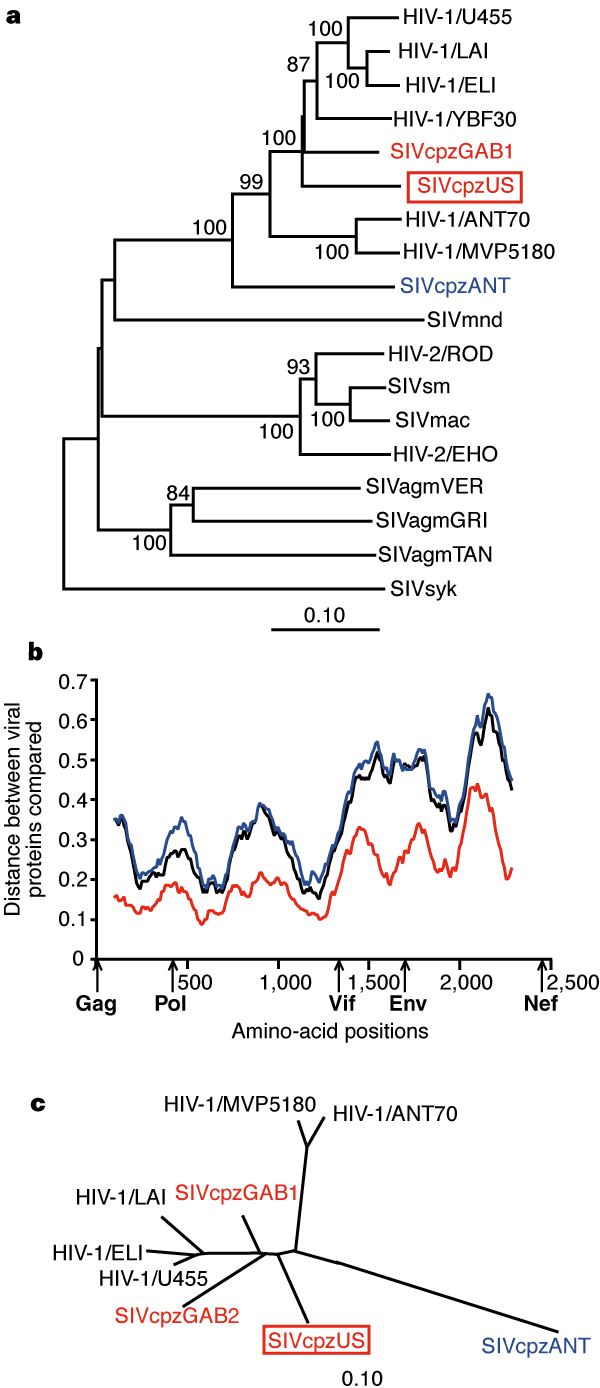
| Figure 1 Phylogenetic analysis of SIVcpzUS. a, Phylogenetic relationship of SIVcpzUS to other primate lentiviruses.
The tree was derived by neighbour-joining analysis27 of full-length
Pol sequences (trees derived by maximum-likelihood methods28
yielded very similar topologies). Horizontal branch lengths are drawn to scale
with the bar indicating 0.1 amino-acid replacements per site. Numbers at each
node indicate the percentage of bootstrap samples (out of 1,000) in which
the cluster to the right is supported (only values >80% are shown). Other
SIVcpz strains closely or more distantly related to SIVcpzUS are shown in
red and blue, respectively. b, Diversity plots of concatenated SIVcpz
protein sequences depicting the proportion of amino-acid sequence differences
between SIVcpzUS and SIVcpzGAB1 (red), SIVcpzUS and SIVcpzANT (blue), and
SIVcpzGAB1 and SIVcpzANT (black), calculated for a window of 200 amino acids
moved in steps of 10 amino acids along the alignment (available as Supplementary
Information). The x-axis shows the amino-acid positions along the alignment.
The positions of Gag, Pol, Vif, Env and Nef regions are shown. The y-axis
denotes the distance between the viral proteins compared (0.1 = 10%
difference). c, Unrooted neighbour-joining tree of partial Pol protein
sequences (distances are drawn to scale).
|