Despite the high importance of membrane proteins in biological function, the number of such structures which have been solved is very small, because of the difficulty in producing good quality crystals. The detergent in which the protein has to be partially solubilised plays a crucial role particularly as it protects the hydrophobic transmembrane surface of the protein during the extraction of the protein from the membrane.
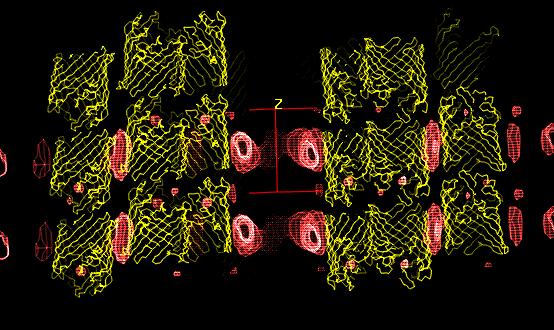
A knowledge of the structure of detergents in protein crystals can yield a better understanding of the interactions between protein and membrane, the detergent playing the same role as the membrane, and of the effect of the detergent in the crystallization process. Using low resolution neutron crystallography (DB21 diffractometer, Institut Laue-Langevin, Grenoble France) combined with contrast variation [1], a well adapted technique for this study of disordered structures, the structure of detergents in two membrane protein crystals, an E. coli porin (P321 crystal [2]) and a Rhodobacter capsulatus porin (R3 crystal [3]), have been solved.
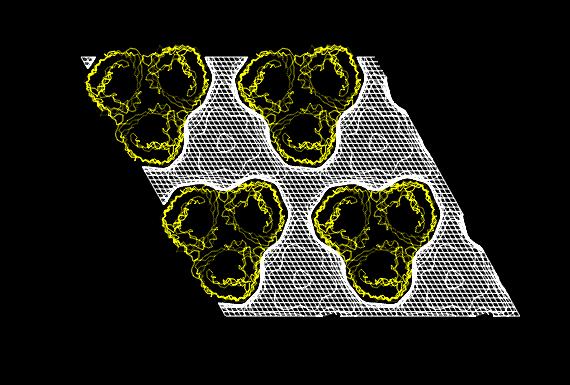
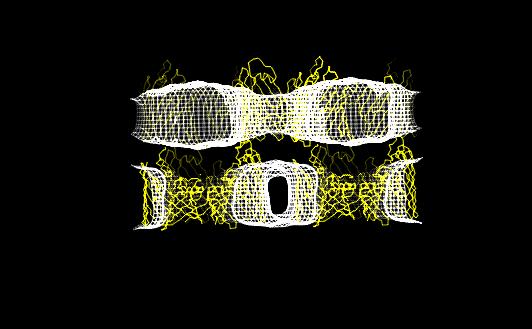
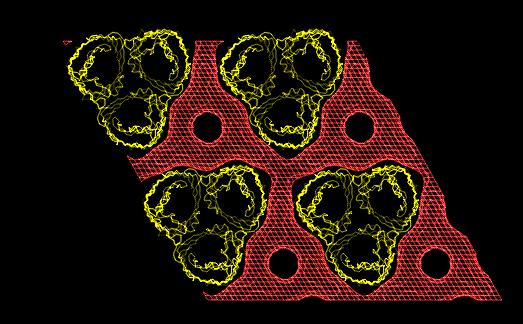
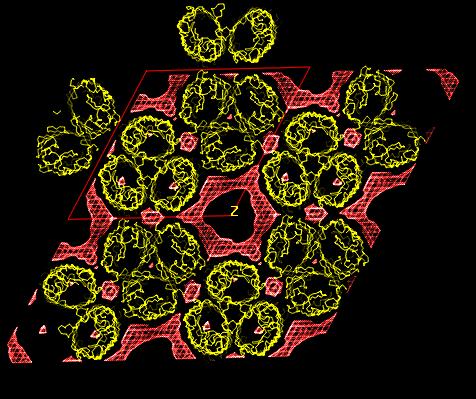
In both studies, the detergent forms micellar
structures around the transmembrane surface of the protein, the micellar
borders are limited by specific interactions with aromatic amino acids
forming two rings playing a role in stabilizing the protein in the membrane.
Theses studies show that the dimensions of the hydrophobic and
hydrophilic moieties of the detergent is a determinant factor in order
to favour protein-protein contacts during the nucleation and the crystal
growth.