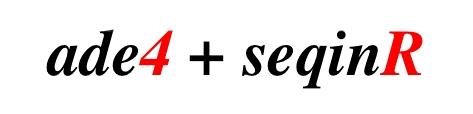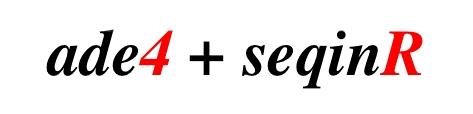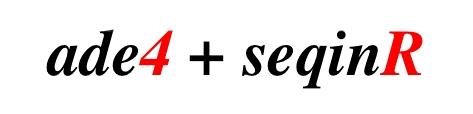



Introduction
External Data Entry
Examples
Introduction
This server allows the use of two packages developped in our
Lab
for the R software:
ade4 and
seqinr.
The documentation of the ade4 package is available
here, and the documentation
of the seqinr package is available
here.
Note that on this server, these two packages are automatically loaded each time R is launched,
so you do not need to use the library(ade4) and library(seqinr) commands
(but using them will not hurt).
To run Rweb just type the code you want
to execute into the text window below and
then click on the submit button.
You will get a new html page with the text output of your code followed
by the graphical output (if any) from your code.
A detailed example of use is here. It shows how you can use
this system to search sequence data banks for gene sequences, compute the codon frequences for these genes,
and perform a correspondence analysis of this data table.
You can try examples from the ade4 package by just clicking the Submit button
with the examples below. Just remove these lines to type your own code.
The computer time for all of this is donated by the
PBIL. Please note that all actions are logged
and that abuse will lead to exclusion of IP addresses.
Examples
Here are a few examples of use of Rweb. You can copy the following code examples and paste them into the text
window at the top of this page. Alternatively, if you are in a more contemplative mood, you can go to
this page and just click and watch.
Example 1
First example using the ade4 library, showing how to do a coinertia analysis :
data(doubs) # load the doubs data set
dudi1 <- dudi.pca(doubs$env, scale = TRUE, scan = FALSE, nf = 3) # first PCA
dudi2 <- dudi.pca(doubs$fish, scale = FALSE, scan = FALSE, nf = 2) # second PCA
coin1 <- coinertia(dudi1,dudi2, scan = FALSE, nf = 2) # coinertia analysis
coin1 # display coinertia analysis results
plot(coin1) # plot coinertia results
r1 <- randtest(coin1) # perform the permutation test
r1 # display it
plot(r1) # plot it
Example 2
Here is an example showing
how you can search sequence data banks for gene sequences (with the seqinR package),
compute the codon frequencies for these genes, and perform a correspondence analysis (CA)
of the frequencies table. This example makes the CA of the codon frequencies in the coding sequences
of Mycoplasma genitalium.
The web page explaining this example is here.
choosebank(bank = "emglib") # chooose the emglib sequence bank
req <- query("liste", "sp=mycoplasma genitalium et t=cds") # search the bank for sequences
seqs <- lapply(liste$req[], getSequence) # retrieve the sequences
tabco <- lapply(seqs, uco) # compute codon frequences
tabco <- as.data.frame(lapply(tabco, as.vector), row.names = names(tabco[[1]])) # make the dataframe
names(tabco) <- liste$req[] # setup codon names
ca <- dudi.coa(tabco, scan = F, nf = 3) # perform CA
s.label(ca$co, clabel = 0, sub = "Genes F1xF2 map") # plot Genes map
s.label(ca$li, sub = "Codons F1xF2 map") # plot codons map
Example 3
Here is another example showing
how you can do a Principal COordinates analysis (PCO) on the distance matrix computed
on a set of aligned sequences in Clustal format. This example also makes use of the seqinR package
(for the dist.alignment function). The web page explaining this example is
here.
mat <- dist.alignment(aln, matrix = "similarity")
dst <- lingoes(mat)
cat <- as.factor(c(1,1,1,1,2,3,3,3,3,3,3,3,4,1,1,1,1,1,1,1,1,3,3,3,3,5,5,5,3))
pco <- dudi.pco(dst, scan = F, nf = 3)
s.label(pco$li, sub = "F1xF2 map")
s.chull(pco$li, cat, optchull=1, add.plot=TRUE, col=c("red","black","green","purple","blue"))
Example 4
Here is a big example, which is in fact the reproduction of the figure from the paper by
Charif, D., Thioulouse, J. Lobry, J.R., Perriere, G. in Bioinformatics (see here).
This example makes intensive use of the seqinR package.
The web page presenting this example is here.
Example 5
Here is a code example showing how an external data set can be used. You must use one of the
three data entry methods to provide data table X, and the PCA of this table will be computed.
X # check that the data table has been read correctly
pca1=dudi.pca(X,scan=F) # do the PCA of this table
scatter(pca1) # PCA biplot
s.corcircle(pca1$co) # PCA correlarion circle
s.label(pca1$li) # PCA row scores
score(pca1) # PCA canonical display : variables = f(principal components)
Example 6
If you don't know how to program in R, here is a short code snippet that
shows you a little bit about what R can do.
x <- rnorm(100) # 100 random numbers from a normal(0,1) distribution
y <- exp(x) + rnorm(100) # an exponential function with error
result <- lsfit(x,y) # regress x on y and store the results
ls.print(result) # print the regression results
plot(x,y) # pretty obvious what this does
abline(result) # add the regression line to the plot
lines(lowess(x,y), col=2) # add a nonparametric regression line (a smoother)
hist(result$residuals) # histogram of the residuals from the regression
© 2004 Jean Thioulouse - Based on the Rweb system by Jeff Banfield.
© 1998 Jeff Banfield
Home Page
<www.math.montana.edu/~umsfjban>
Email
<jeff@math.montana.edu>
Last Modified: 10-Nov-2008