Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Epsilonproteobacteria; Campylobacterales; Helicobacteraceae; Helicobacter.
Genome size (bp): 1667867
Number of genes 1572
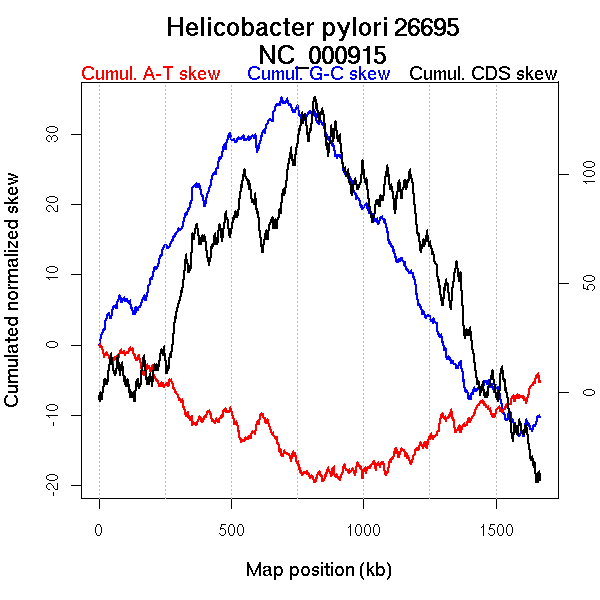
Oriloc predictions: Origin 0 kb Terminus 813 kb
Worning et al., 2006: Origin 1655 kb Terminus 768 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 1271.097 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 1608.31 kb
Consensus predictions: Origin 0 kb Terminus 813 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 375 | NA | 710 |
| GC-skew reverse | 1070 | NA | 790 |
| AT-skew forward | 667 | NA | 1383 |
| AT-skew reverse | 984 | NA | 570 |
| 1355 | NA | 1317 |
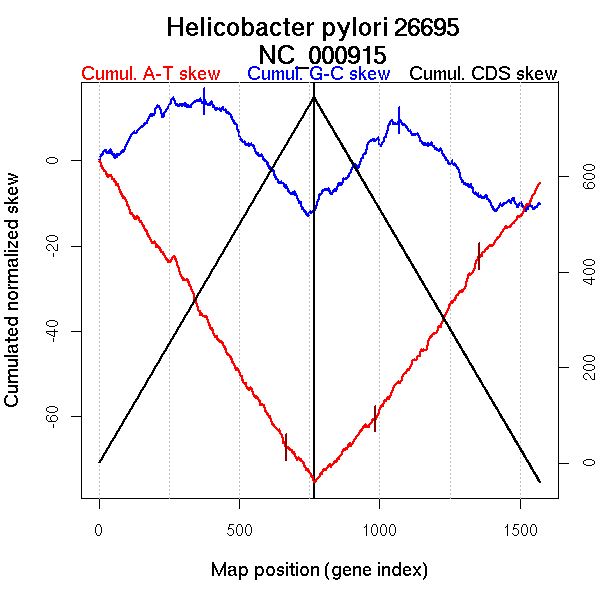
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 375 (709.967 kb) | leading | 0.042 |
| 376 (710.692 kb) | 766 (1667.8 kb) | NA | -0.075 | |
| GC-skew reverse | 767 (0 kb) | 1070 (790.2695 kb) | leading | 0.068 |
| 1071 (791.489 kb) | 1572 (1667.8 kb) | lagging | -0.045 | |
| AT-skew forward | 1 (0 kb) | 667 (1382.6155 kb) | NA | -0.1 |
| 668 (1384.5525 kb) | 766 (1667.8 kb) | lagging | -0.078 | |
| AT-skew reverse | 767 (0 kb) | 984 (569.7705 kb) | leading | 0.07 |
| 985 (571.799 kb) | 1355 (1317.115 kb) | NA | 0.098 | |
| 1356 (1319.218 kb) | 1572 (1667.8 kb) | lagging | 0.076 |