Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Epsilonproteobacteria; Campylobacterales; Helicobacteraceae; Helicobacter.
Genome size (bp): 1643831
Number of genes 1477
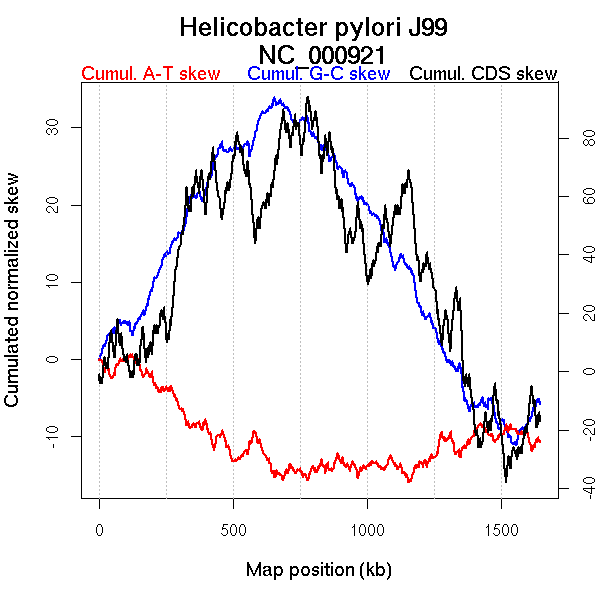
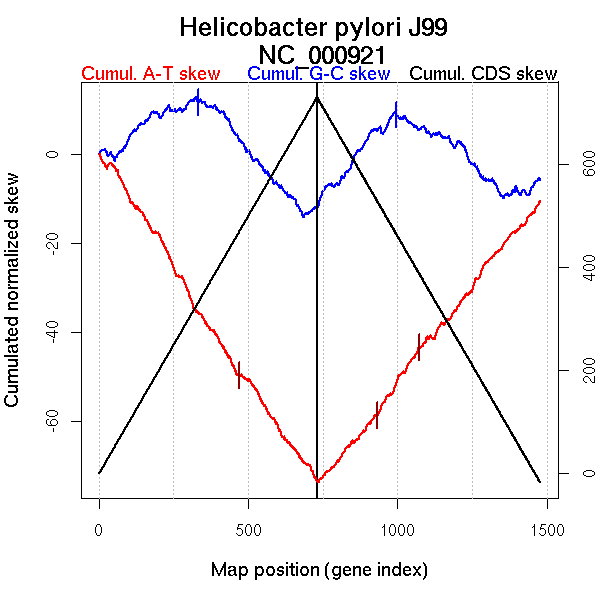
Oriloc predictions: Origin 0 kb Terminus 685 kb
Worning et al., 2006: Origin 1557 kb Terminus 685 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 120.75 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 1558.48 kb
Consensus predictions: Origin 0 kb Terminus 685 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 331 | NA | 663 |
| GC-skew reverse | 995 | NA | 693 |
| AT-skew forward | 468 | NA | 1013 |
| AT-skew reverse | 933 | NA | 536 |
| 1072 | NA | 850 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 331 (663.138 kb) | leading | 0.043 |
| 332 (665.9005 kb) | 730 (1643.586 kb) | lagging | -0.074 | |
| GC-skew reverse | 731 (0 kb) | 995 (693.289 kb) | leading | 0.078 |
| 996 (695.155 kb) | 1477 (1643.586 kb) | lagging | -0.043 | |
| AT-skew forward | 1 (0 kb) | 468 (1013.4015 kb) | NA | -0.109 |
| 469 (1016.4615 kb) | 730 (1643.586 kb) | lagging | -0.096 | |
| AT-skew reverse | 731 (0 kb) | 933 (536.4565 kb) | leading | 0.077 |
| 934 (537.257 kb) | 1072 (849.739 kb) | NA | 0.104 | |
| 1073 (854.855 kb) | 1477 (1643.586 kb) | lagging | 0.084 |
More G than C on the leading strand for replication.