Rearranged Oriloc output example: Previous Index Next
Bacteria; Actinobacteria; Actinobacteridae; Actinomycetales; Corynebacterineae; Mycobacteriaceae; Mycobacterium; Mycobacterium tuberculosis complex.
Genome size (bp): 4403837
Number of genes 4189
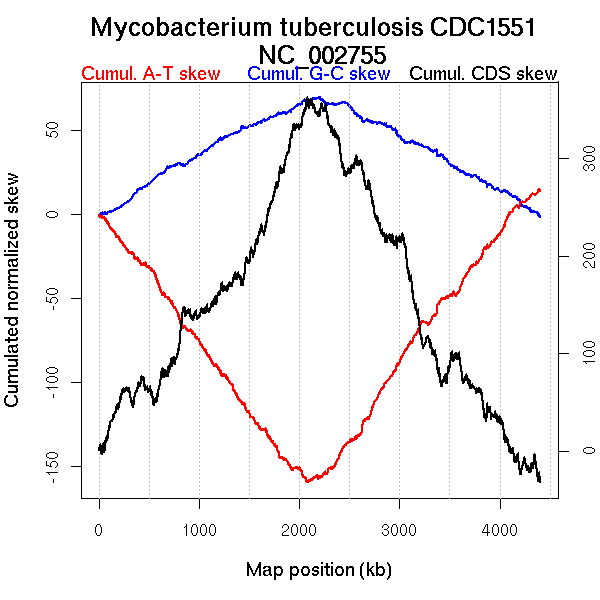
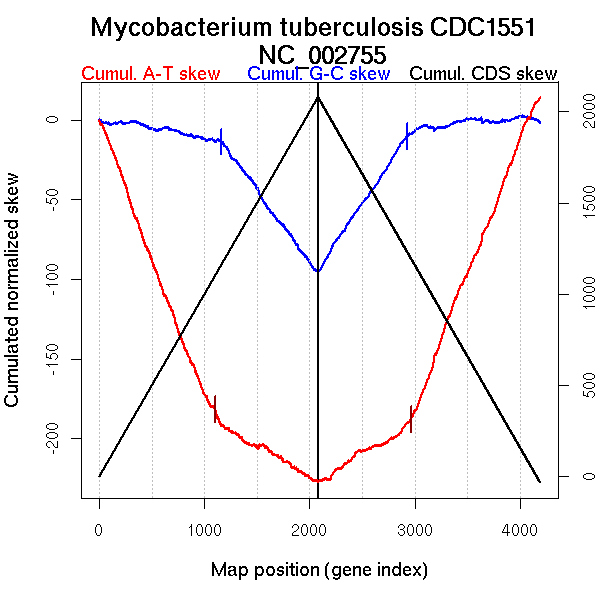
Oriloc predictions: Origin 0 kb Terminus 2184 kb
Worning et al., 2006: Origin 2 kb Terminus 2125 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 60.113 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.76 kb
Consensus predictions: Origin 0 kb Terminus 2184 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 1162 | NA | 2109 |
| GC-skew reverse | 2925 | NA | 2183 |
| 3630 | NA | 3365 | |
| 3799 | NA | 3674 | |
| AT-skew forward | 816 | NA | 1510 |
| 1167 | NA | 2122 | |
| AT-skew reverse | 2960 | NA | 2244 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 1162 (2109.0425 kb) | leading | -0.012 |
| 1163 (2113.631 kb) | 2079 (4403.468 kb) | lagging | -0.091 | |
| GC-skew reverse | 2080 (0 kb) | 2925 (2183.3925 kb) | leading | 0.099 |
| 2926 (2187.9335 kb) | 3630 (3365.2175 kb) | lagging | 0.013 | |
| 3631 (3367.385 kb) | 3799 (3673.9425 kb) | lagging | -0.009 | |
| 3800 (3678.907 kb) | 4189 (4403.468 kb) | lagging | 0.003 | |
| AT-skew forward | 1 (0 kb) | 816 (1509.79 kb) | leading | -0.179 |
| 817 (1510.5815 kb) | 1167 (2122.3015 kb) | leading | -0.139 | |
| 1168 (2123.081 kb) | 2079 (4403.468 kb) | lagging | -0.038 | |
| AT-skew reverse | 2080 (0 kb) | 2960 (2243.9525 kb) | leading | 0.041 |
| 2961 (2247.109 kb) | 4189 (4403.468 kb) | lagging | 0.168 |
More G than C on the leading strand for replication.