Rearranged Oriloc output example: Previous Index Next
Bacteria; Actinobacteria; Actinobacteridae; Actinomycetales; Corynebacterineae; Mycobacteriaceae; Mycobacterium.
Genome size (bp): 3268203
Number of genes 1605
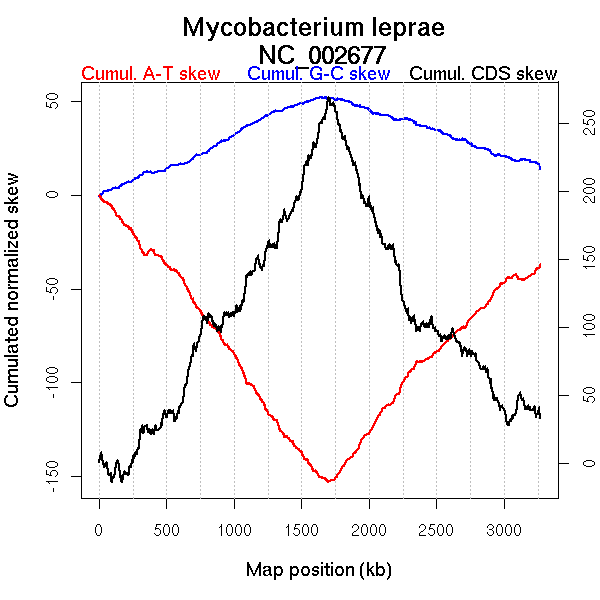
Oriloc predictions: Origin 0 kb Terminus 1706 kb
Worning et al., 2006: Origin 2 kb Terminus 1700 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 17.03 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.78 kb
Consensus predictions: Origin 0 kb Terminus 1706 kb
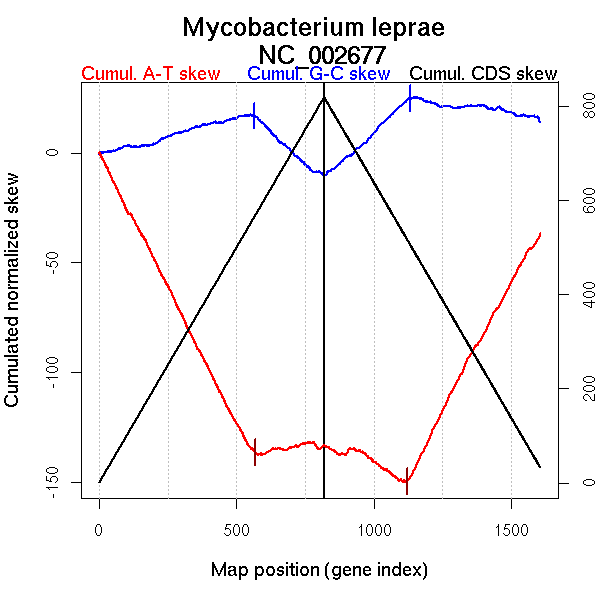
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 563 | NA | 1681 |
| GC-skew reverse | 1134 | NA | 1740 |
| 1252 | NA | 2053 | |
| 1377 | NA | 2383 | |
| AT-skew forward | 569 | NA | 1692 |
| 762 | NA | 2971 | |
| AT-skew reverse | 1121 | NA | 1657 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 563 (1680.611 kb) | leading | 0.034 |
| 564 (1682.086 kb) | 819 (3267.653 kb) | lagging | -0.113 | |
| GC-skew reverse | 820 (0 kb) | 1134 (1740.069 kb) | leading | 0.118 |
| 1135 (1743.831 kb) | 1252 (2053.3055 kb) | lagging | -0.04 | |
| 1253 (2054.013 kb) | 1377 (2382.666 kb) | lagging | 0.004 | |
| 1378 (2390.552 kb) | 1605 (3267.653 kb) | lagging | -0.028 | |
| AT-skew forward | 1 (0 kb) | 569 (1692.1175 kb) | leading | -0.246 |
| 570 (1693.097 kb) | 762 (2970.906 kb) | lagging | 0.027 | |
| 763 (2974.6035 kb) | 819 (3267.653 kb) | lagging | -0.051 | |
| AT-skew reverse | 820 (0 kb) | 1121 (1657.49 kb) | leading | -0.055 |
| 1122 (1666.688 kb) | 1605 (3267.653 kb) | lagging | 0.236 |
More G than C on the leading strand for replication.