Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Deltaproteobacteria; Desulfovibrionales; Desulfovibrionaceae; Desulfovibrio.
Genome size (bp): 3570858
Number of genes 3378
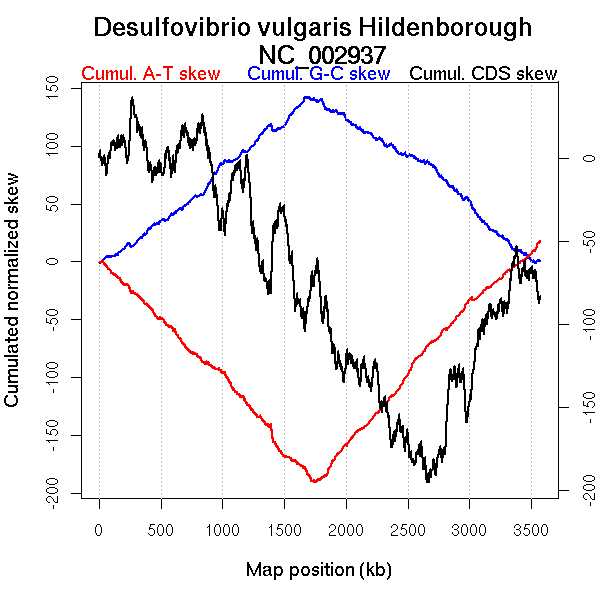
Oriloc predictions: Origin 0 kb Terminus 1666 kb
Worning et al., 2006: Origin 9 kb Terminus 1747 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 2271.812 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.81 kb, 2348.45 kb
Consensus predictions: Origin 0 kb Terminus 1666 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 783 | NA | 1722 |
| 1242 | NA | 2787 | |
| 1287 | NA | 2830 | |
| GC-skew reverse | 2520 | NA | 1770 |
| AT-skew forward | 375 | NA | 823 |
| 609 | NA | 1393 | |
| 644 | NA | 1412 | |
| 795 | NA | 1746 | |
| 961 | NA | 2137 | |
| AT-skew reverse | 2540 | NA | 1790 |
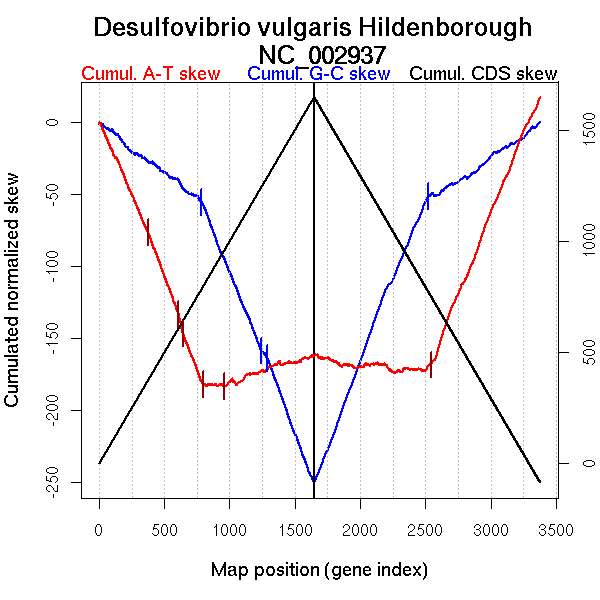
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 783 (1721.9025 kb) | leading | -0.069 |
| 784 (1724.112 kb) | 1242 (2787.0295 kb) | lagging | -0.224 | |
| 1243 (2787.9375 kb) | 1287 (2829.9325 kb) | lagging | -0.099 | |
| 1288 (2830.997 kb) | 1647 (3570.714 kb) | lagging | -0.247 | |
| GC-skew reverse | 1648 (0 kb) | 2520 (1769.8165 kb) | leading | 0.232 |
| 2521 (1770.351 kb) | 3378 (3570.714 kb) | lagging | 0.062 | |
| AT-skew forward | 1 (0 kb) | 375 (822.632 kb) | leading | -0.206 |
| 376 (825.193 kb) | 609 (1392.81 kb) | leading | -0.242 | |
| 610 (1393.226 kb) | 644 (1411.882 kb) | leading | -0.403 | |
| 645 (1413.2 kb) | 795 (1745.8345 kb) | NA | -0.232 | |
| 796 (1748.2385 kb) | 961 (2136.605 kb) | lagging | -0.002 | |
| 962 (2136.799 kb) | 1647 (3570.714 kb) | lagging | 0.03 | |
| AT-skew reverse | 1648 (0 kb) | 2540 (1790.389 kb) | leading | -0.009 |
| 2541 (1791.715 kb) | 3378 (3570.714 kb) | lagging | 0.225 |
More G than C on the leading strand for replication.