Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae; Yersinia.
Genome size (bp): 4653728
Number of genes 3870
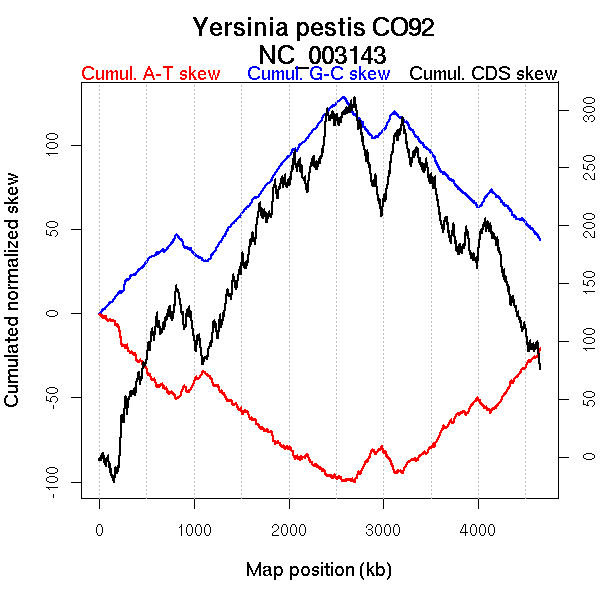
Oriloc predictions: Origin 0 kb Terminus 2580 kb
Worning et al., 2006: Origin 0 kb Terminus 2547 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 185.682 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 3427.28 kb, 4619.03 kb
Consensus predictions: Origin 0 kb Terminus 2580 kb
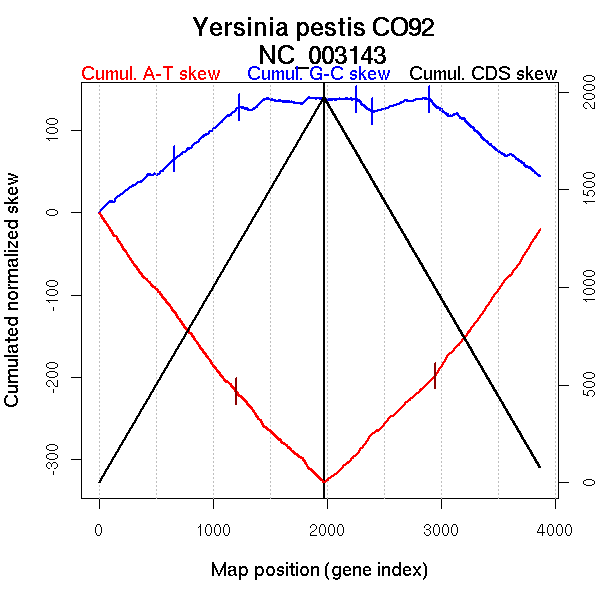
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 1216 | NA | 2557 |
| GC-skew reverse | 2891 | NA | 2546 |
| AT-skew forward | 288 | NA | 537 |
| 1092 | NA | 2326 | |
| 1451 | NA | 3120 | |
| AT-skew reverse | 2949 | NA | 2698 |
| 3669 | NA | 4252 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 1216 (2557.412 kb) | leading | 0.1 |
| 1217 (2557.715 kb) | 1973 (4653.617 kb) | lagging | 0.015 | |
| GC-skew reverse | 1974 (0 kb) | 2891 (2545.703 kb) | leading | -0.002 |
| 2892 (2547.9225 kb) | 3870 (4653.617 kb) | lagging | -0.096 | |
| AT-skew forward | 1 (0 kb) | 288 (537.009 kb) | leading | -0.203 |
| 289 (538.866 kb) | 1092 (2326.1355 kb) | leading | -0.178 | |
| 1093 (2329.317 kb) | 1451 (3119.884 kb) | NA | -0.161 | |
| 1452 (3121.9475 kb) | 1973 (4653.617 kb) | lagging | -0.13 | |
| AT-skew reverse | 1974 (0 kb) | 2949 (2697.6315 kb) | leading | 0.137 |
| 2950 (2698.6515 kb) | 3669 (4252.1505 kb) | lagging | 0.185 | |
| 3670 (4254.1315 kb) | 3870 (4653.617 kb) | lagging | 0.224 |
More G than C on the leading strand for replication.