Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae; Yersinia.
Genome size (bp): 4600755
Number of genes 4086
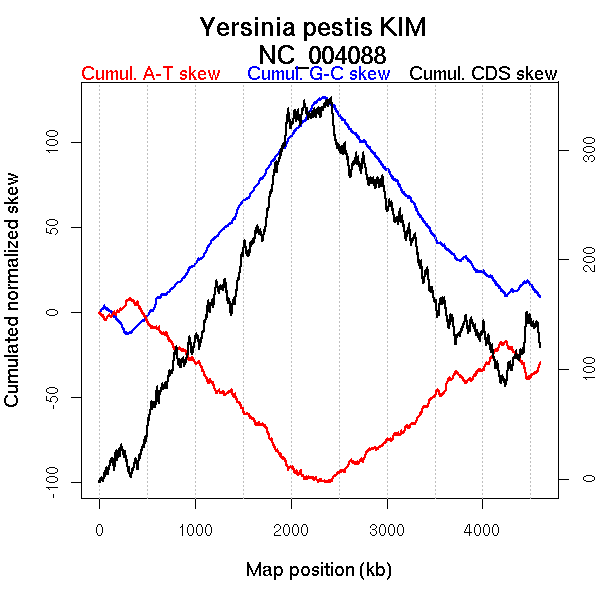
Oriloc predictions: Origin 318 kb Terminus 2348 kb
Worning et al., 2006: Origin 333 kb Terminus 2324 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 4408.751 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 1567.37 kb, 4565.82 kb
Consensus predictions: Origin 318 kb Terminus 2348 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 1202 | NA | 2318 |
| GC-skew reverse | 2968 | NA | 2320 |
| AT-skew forward | 155 | NA | 337 |
| 1134 | NA | 2166 | |
| 1916 | NA | 4216 | |
| AT-skew reverse | 2229 | NA | 293 |
| 3058 | NA | 2484 |
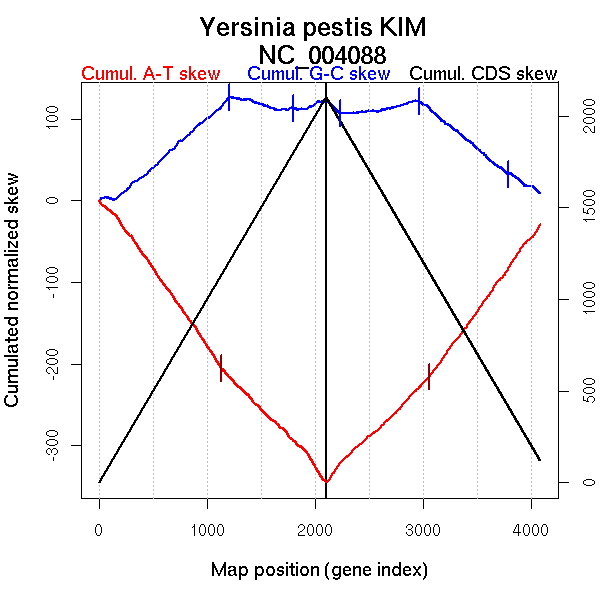
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 1202 (2317.6175 kb) | leading | 0.111 |
| 1203 (2319.9655 kb) | 2103 (4600.476 kb) | lagging | -0.011 | |
| GC-skew reverse | 2104 (0 kb) | 2968 (2319.899 kb) | NA | 0.011 |
| 2969 (2322.1105 kb) | 4086 (4600.476 kb) | lagging | -0.103 | |
| AT-skew forward | 1 (0 kb) | 155 (337.067 kb) | lagging | -0.106 |
| 156 (339.9695 kb) | 1134 (2165.7615 kb) | leading | -0.189 | |
| 1135 (2167.21 kb) | 1916 (4215.7175 kb) | lagging | -0.13 | |
| 1917 (4218.694 kb) | 2103 (4600.476 kb) | lagging | -0.213 | |
| AT-skew reverse | 2104 (0 kb) | 2229 (293.0815 kb) | lagging | 0.199 |
| 2230 (294.723 kb) | 3058 (2483.9075 kb) | leading | 0.127 | |
| 3059 (2484.838 kb) | 4086 (4600.476 kb) | lagging | 0.182 |
More G than C on the leading strand for replication - for forward encoded genes.