Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae; Buchnera.
Genome size (bp): 641454
Number of genes 546
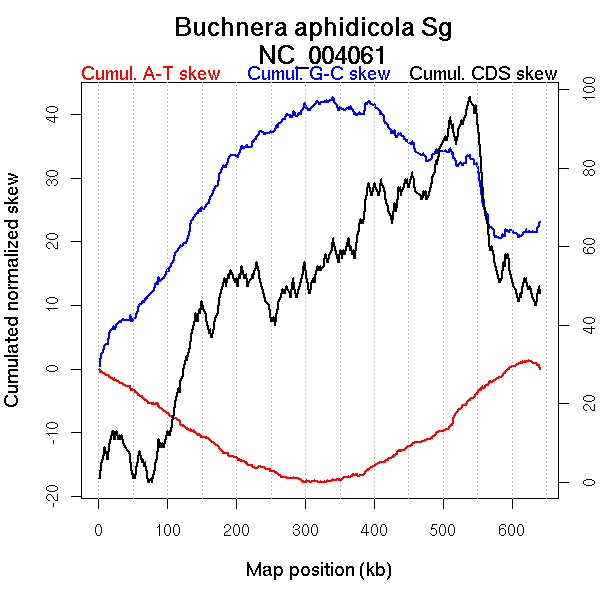
Oriloc predictions: Origin 0 kb Terminus 329 kb
Worning et al., 2006: Origin 628 kb Terminus 331 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 121.963 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 13.08 kb
Consensus predictions: Origin 0 kb Terminus 329 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 154 | NA | 308 |
| 260 | NA | 522 | |
| GC-skew reverse | 399 | NA | 304 |
| 516 | NA | 576 | |
| AT-skew forward | 169 | NA | 339 |
| AT-skew reverse | 397 | NA | 302 |
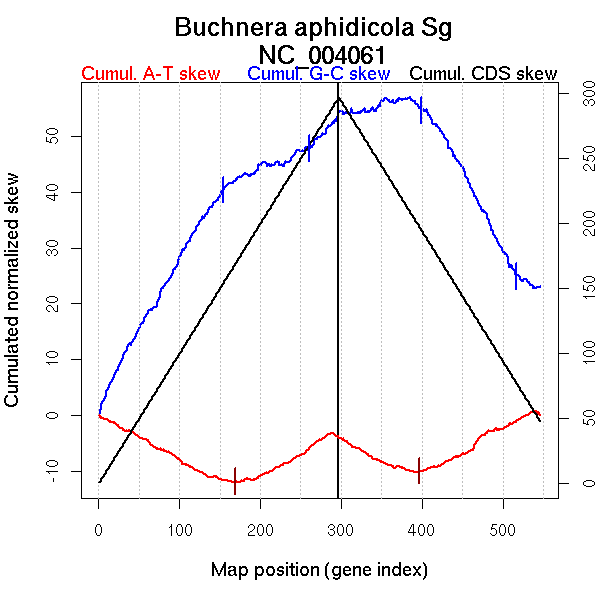
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 154 (308.427 kb) | leading | 0.255 |
| 155 (309.5565 kb) | 260 (521.7875 kb) | lagging | 0.062 | |
| 261 (523.2085 kb) | 297 (641.021 kb) | lagging | 0.162 | |
| GC-skew reverse | 298 (0 kb) | 399 (304.2175 kb) | leading | 0.027 |
| 400 (305.3405 kb) | 516 (576.395 kb) | lagging | -0.269 | |
| 517 (577.5765 kb) | 546 (641.021 kb) | lagging | -0.083 | |
| AT-skew forward | 1 (0 kb) | 169 (338.815 kb) | leading | -0.076 |
| 170 (339.9225 kb) | 297 (641.021 kb) | lagging | 0.078 | |
| AT-skew reverse | 298 (0 kb) | 397 (302.0995 kb) | leading | -0.064 |
| 398 (303.12 kb) | 546 (641.021 kb) | lagging | 0.081 |
More G than C on the leading strand for replication.