Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae; Buchnera.
Genome size (bp): 615980
Number of genes 504
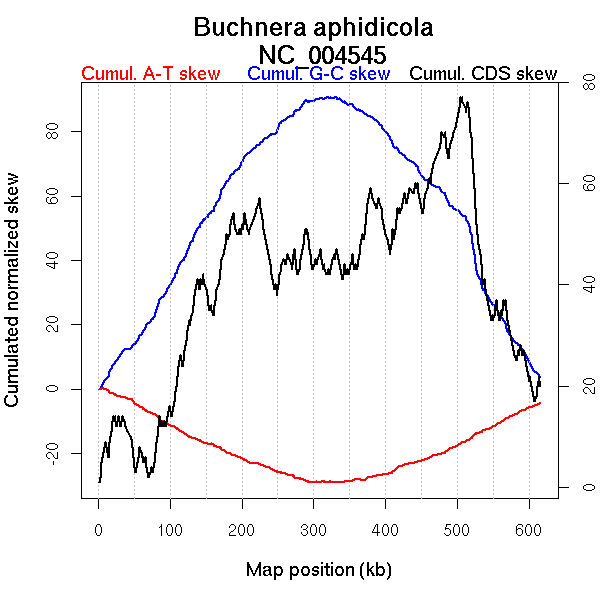
Oriloc predictions: Origin 4 kb Terminus 336 kb
Worning et al., 2006: Origin 3 kb Terminus 320 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 86.816 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 13.8 kb
Consensus predictions: Origin 0 kb Terminus 336 kb
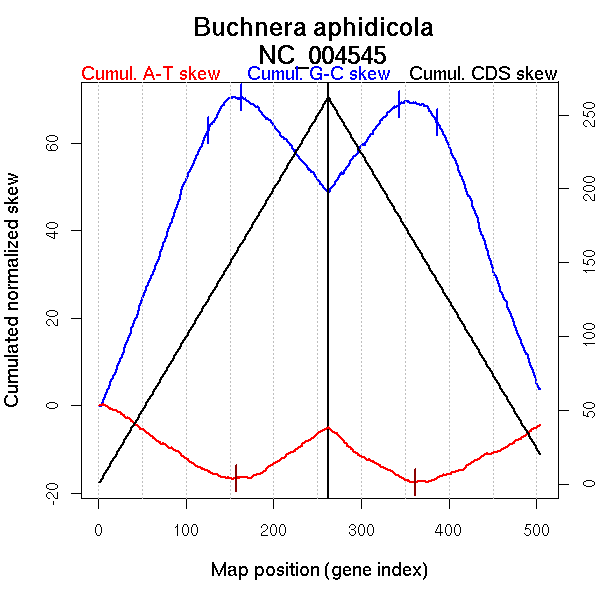
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 125 | 0.02333 | 256 |
| 163 | 0.00333 | 354 | |
| GC-skew reverse | 343 | 0.04333 | 255 |
| 387 | 0.00333 | 384 | |
| AT-skew forward | 157 | 0 | 342 |
| AT-skew reverse | 361 | 0 | 298 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 125 (255.741 kb) | leading | 0.529 |
| 126 (260.2555 kb) | 163 (354.121 kb) | NA | 0.197 | |
| 164 (355.9595 kb) | 262 (615.98 kb) | lagging | -0.231 | |
| GC-skew reverse | 263 (0 kb) | 343 (254.7085 kb) | leading | 0.258 |
| 344 (262.909 kb) | 387 (383.894 kb) | NA | -0.092 | |
| 388 (385.1935 kb) | 504 (615.98 kb) | lagging | -0.542 | |
| AT-skew forward | 1 (0 kb) | 157 (342.212 kb) | leading | -0.123 |
| 158 (342.4955 kb) | 262 (615.98 kb) | lagging | 0.123 | |
| AT-skew reverse | 263 (0 kb) | 361 (297.918 kb) | leading | -0.126 |
| 362 (301.2495 kb) | 504 (615.98 kb) | lagging | 0.095 |
More G than C on the leading strand for replication.