Rearranged Oriloc output example: Previous Index Next
Bacteria; Spirochaetes; Spirochaetales; Leptospiraceae; Leptospira.
Genome size (bp): 358943
Number of genes 367
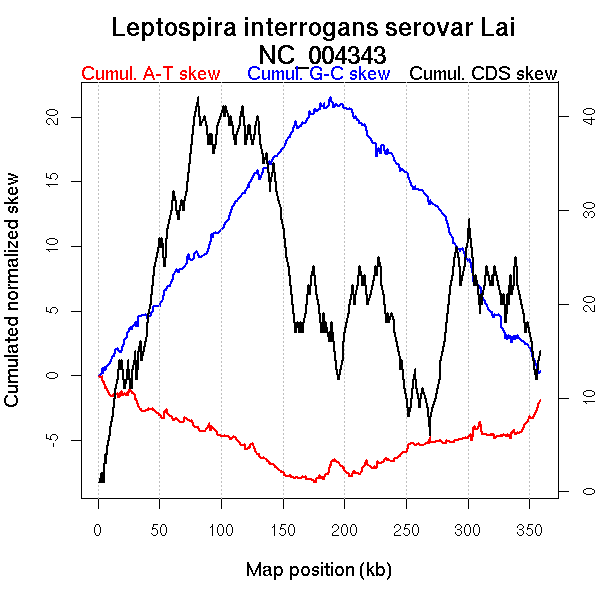
Oriloc predictions: Origin 0 kb Terminus 205 kb
Worning et al., 2006: Origin 357 kb Terminus 205 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 36.073 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): NA
Consensus predictions: Origin 0 kb Terminus 205 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 108 | NA | 197 |
| GC-skew reverse | 280 | NA | 189 |
| AT-skew forward | 104 | NA | 186 |
| AT-skew reverse | 234 | NA | 119 |
| 266 | NA | 164 | |
| 335 | NA | 308 |
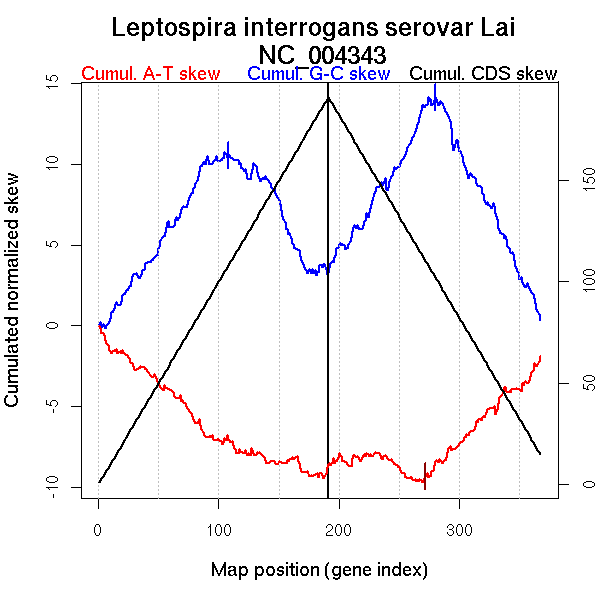
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 108 (197.1905 kb) | leading | 0.111 |
| 109 (198.324 kb) | 191 (358.928 kb) | lagging | -0.107 | |
| GC-skew reverse | 192 (0 kb) | 280 (189.115 kb) | leading | 0.121 |
| 281 (190.3705 kb) | 367 (358.928 kb) | lagging | -0.154 | |
| AT-skew forward | 1 (0 kb) | 104 (185.9605 kb) | leading | -0.067 |
| 105 (188.284 kb) | 191 (358.928 kb) | lagging | -0.023 | |
| AT-skew reverse | 192 (0 kb) | 234 (119.4255 kb) | leading | 0.013 |
| 235 (121.165 kb) | 266 (164.3245 kb) | leading | -0.058 | |
| 267 (168.265 kb) | 335 (307.8385 kb) | NA | 0.077 | |
| 336 (308.6945 kb) | 367 (358.928 kb) | lagging | 0.063 |
More G than C on the leading strand for replication.