Rearranged Oriloc output example: Previous Index Next
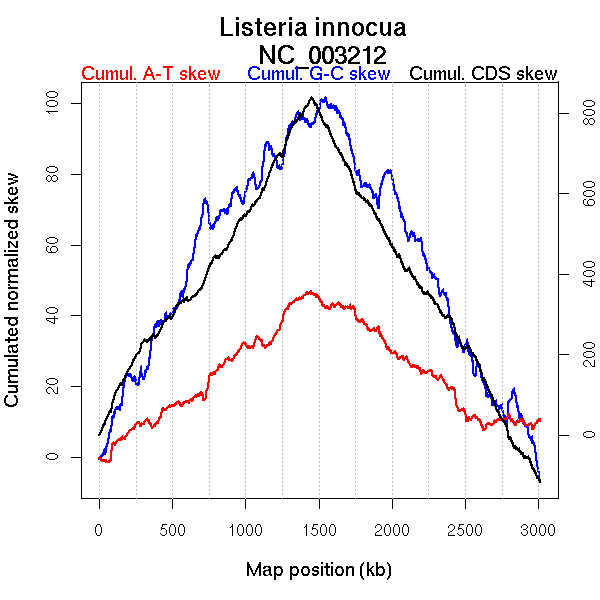
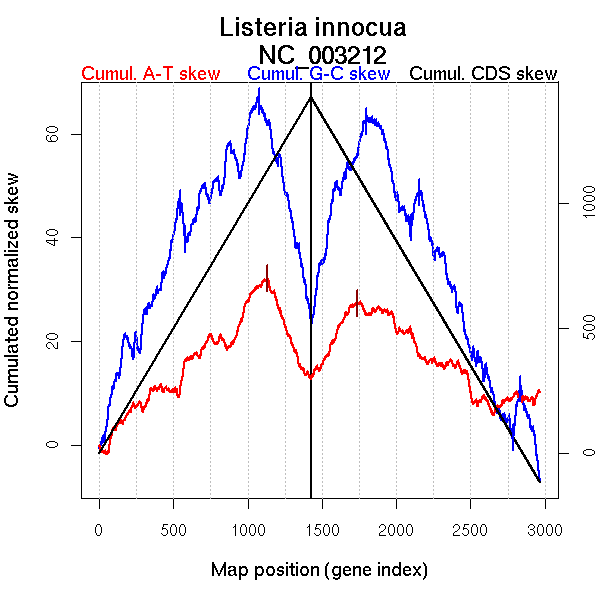
Bacteria; Firmicutes; Bacillales; Listeriaceae; Listeria.
Genome size (bp): 3011208
Number of genes 2967
Oriloc predictions: Origin 0 kb Terminus 1419 kb
Worning et al., 2006: Origin 5 kb Terminus 1445 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 1.725 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 1 kb, 1603.59 kb
Consensus predictions: Origin 0 kb Terminus 1419 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 544 | 0 | 728 |
| 582 | 0 | 768 | |
| 1074 | 0.00667 | 1366 | |
| GC-skew reverse | 1796 | 0.02933 | 1533 |
| 2100 | 0.00933 | 1914 | |
| 2151 | 0.012 | 1973 | |
| 2788 | 0.00667 | 2787 | |
| 2830 | 0.00533 | 2825 | |
| AT-skew forward | 1132 | 0.00667 | 1442 |
| AT-skew reverse | 1735 | 0 | 1464 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 544 (727.876 kb) | leading | 0.074 |
| 545 (728.3035 kb) | 582 (767.6665 kb) | leading | -0.231 | |
| 583 (768.346 kb) | 1074 (1365.547 kb) | leading | 0.047 | |
| 1075 (1366.0085 kb) | 1425 (3011.02 kb) | lagging | -0.119 | |
| GC-skew reverse | 1426 (0 kb) | 1796 (1532.541 kb) | NA | 0.097 |
| 1797 (1533.204 kb) | 2100 (1913.931 kb) | lagging | -0.084 | |
| 2101 (1914.456 kb) | 2151 (1973.41 kb) | lagging | 0.141 | |
| 2152 (1974.6145 kb) | 2788 (2787.29 kb) | lagging | -0.074 | |
| 2789 (2788.056 kb) | 2830 (2824.8665 kb) | lagging | 0.196 | |
| 2831 (2825.541 kb) | 2967 (3011.02 kb) | lagging | -0.12 | |
| AT-skew forward | 1 (0 kb) | 1132 (1442.3505 kb) | leading | 0.027 |
| 1133 (1444.8335 kb) | 1425 (3011.02 kb) | lagging | -0.07 | |
| AT-skew reverse | 1426 (0 kb) | 1735 (1464.4445 kb) | leading | 0.052 |
| 1736 (1465.6445 kb) | 2967 (3011.02 kb) | lagging | -0.018 |
More G than C on the leading strand for replication - for forward encoded genes.