Rearranged Oriloc output example: Previous Index Next
Bacteria; Firmicutes; Lactobacillales; Streptococcaceae; Streptococcus.
Genome size (bp): 2030921
Number of genes 1960
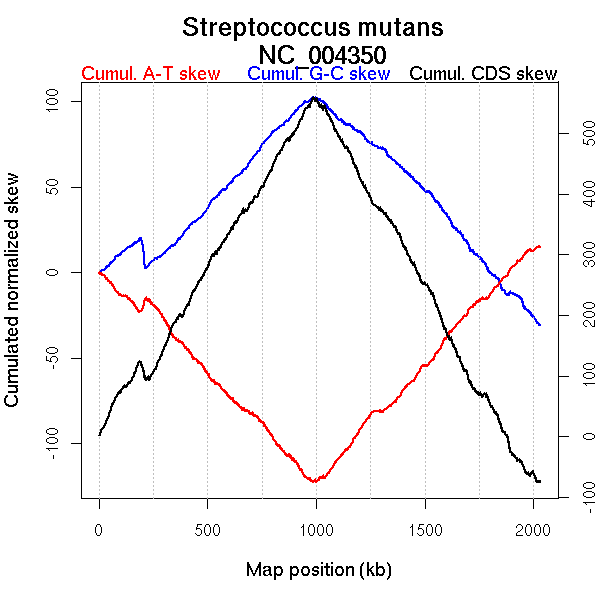
Oriloc predictions: Origin 0 kb Terminus 1001 kb
Worning et al., 2006: Origin 2022 kb Terminus 1000 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 0.092 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.87 kb
Consensus predictions: Origin 0 kb Terminus 1001 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 747 | NA | 981 |
| GC-skew reverse | 1003 | NA | 227 |
| 1145 | NA | 1002 | |
| 1315 | NA | 1209 | |
| AT-skew forward | 45 | NA | 54 |
| 566 | NA | 745 | |
| 729 | NA | 961 | |
| AT-skew reverse | 1075 | NA | 663 |
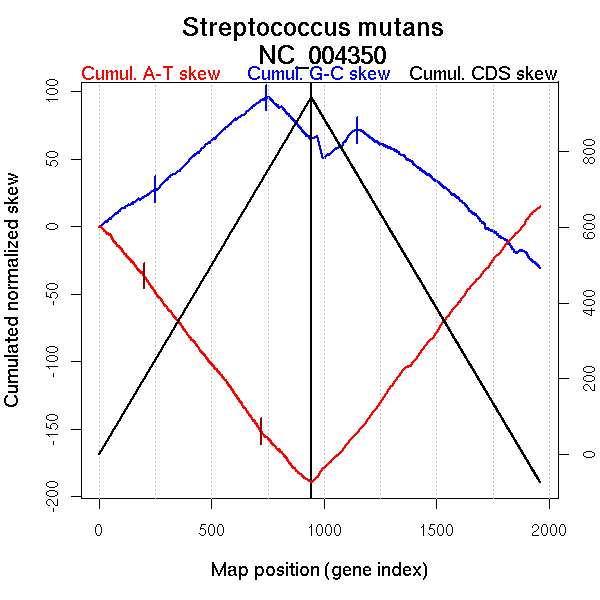
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 747 (980.888 kb) | leading | 0.133 |
| 748 (981.315 kb) | 943 (2030.91 kb) | lagging | -0.164 | |
| GC-skew reverse | 944 (0 kb) | 1003 (227.451 kb) | leading | -0.319 |
| 1004 (228.43 kb) | 1145 (1001.8985 kb) | leading | 0.153 | |
| 1146 (1002.5775 kb) | 1315 (1208.7065 kb) | lagging | -0.097 | |
| 1316 (1209.872 kb) | 1960 (2030.91 kb) | lagging | -0.138 | |
| AT-skew forward | 1 (0 kb) | 45 (54.342 kb) | leading | -0.148 |
| 46 (55.4265 kb) | 566 (744.56 kb) | leading | -0.212 | |
| 567 (745.314 kb) | 729 (961.1215 kb) | leading | -0.239 | |
| 730 (963.1465 kb) | 943 (2030.91 kb) | lagging | -0.177 | |
| AT-skew reverse | 944 (0 kb) | 1075 (662.5125 kb) | leading | 0.176 |
| 1076 (663.0685 kb) | 1960 (2030.91 kb) | lagging | 0.208 |
More G than C on the leading strand for replication.