Rearranged Oriloc output example: Previous Index Next
Bacteria; Firmicutes; Lactobacillales; Streptococcaceae; Streptococcus.
Genome size (bp): 2038615
Number of genes 2043
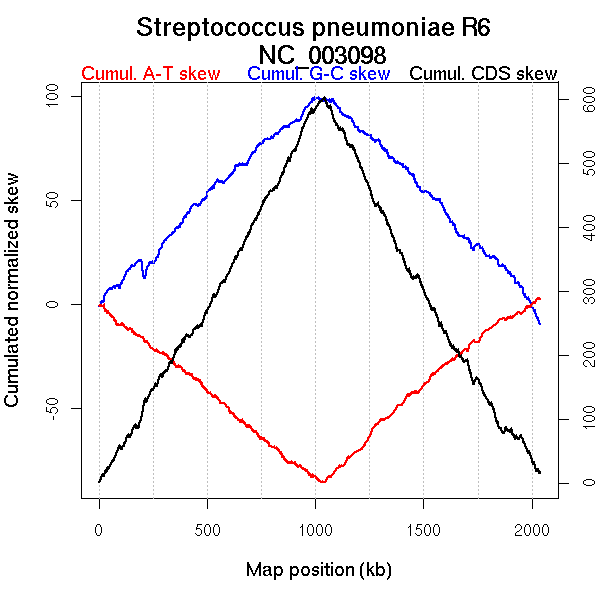
Oriloc predictions: Origin 0 kb Terminus 1040 kb
Worning et al., 2006: Origin 2037 kb Terminus 1041 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 2038.503 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.68 kb, 1537.67 kb
Consensus predictions: Origin 0 kb Terminus 1040 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 810 | NA | 1010 |
| GC-skew reverse | 1255 | NA | 1046 |
| 1713 | NA | 1587 | |
| 1974 | NA | 1952 | |
| AT-skew forward | 62 | NA | 84 |
| 173 | NA | 210 | |
| 694 | NA | 873 | |
| AT-skew reverse | 1172 | NA | 658 |
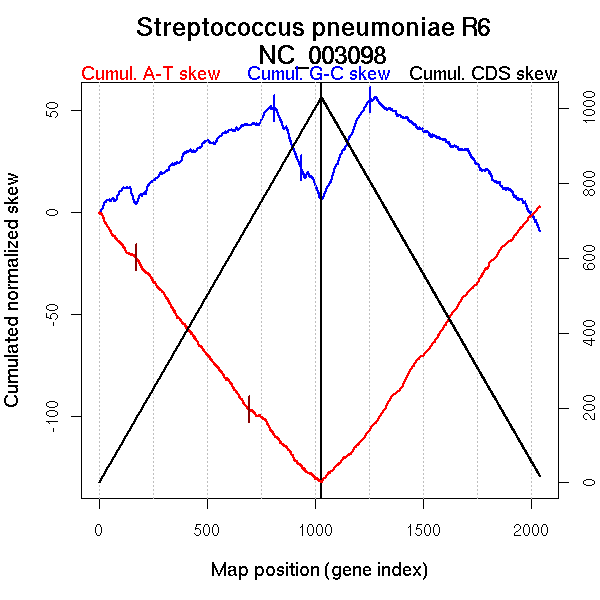
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 810 (1010.034 kb) | leading | 0.062 |
| 811 (1012.1895 kb) | 1030 (2038.4 kb) | lagging | -0.206 | |
| GC-skew reverse | 1031 (0 kb) | 1255 (1046.2735 kb) | leading | 0.226 |
| 1256 (1047.535 kb) | 1713 (1587.192 kb) | lagging | -0.06 | |
| 1714 (1587.694 kb) | 1974 (1951.866 kb) | lagging | -0.09 | |
| 1975 (1954.101 kb) | 2043 (2038.4 kb) | lagging | -0.185 | |
| AT-skew forward | 1 (0 kb) | 62 (83.753 kb) | leading | -0.193 |
| 63 (84.533 kb) | 173 (210.4285 kb) | leading | -0.108 | |
| 174 (210.8785 kb) | 694 (872.628 kb) | leading | -0.141 | |
| 695 (874.4405 kb) | 1030 (2038.4 kb) | NA | -0.117 | |
| AT-skew reverse | 1031 (0 kb) | 1172 (657.6575 kb) | leading | 0.102 |
| 1173 (658.338 kb) | 2043 (2038.4 kb) | lagging | 0.141 |
More G than C on the leading strand for replication.