Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Bradyrhizobiaceae; Bradyrhizobium.
Genome size (bp): 9105828
Number of genes 8317
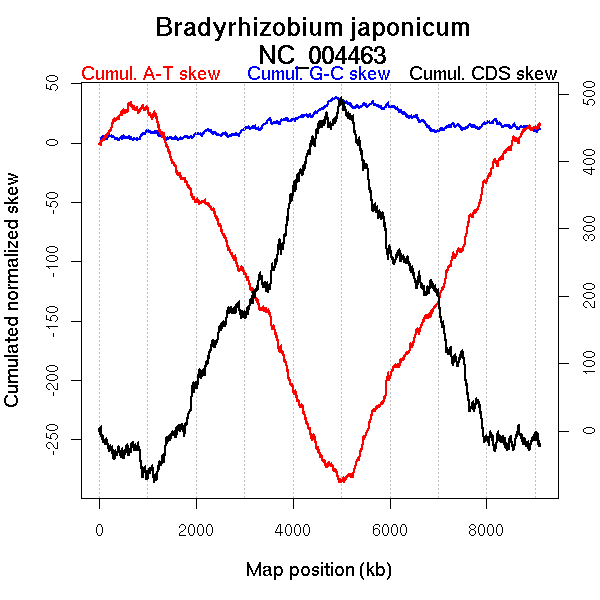
Oriloc predictions: Origin 684 kb Terminus 4926 kb
Worning et al., 2006: Origin 328 kb Terminus 5002 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 68.858 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 894.42 kb
Consensus predictions: Origin 684 kb Terminus 4926 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 757 | 0 | 1646 |
| 1135 | 0.00667 | 2319 | |
| 2482 | 0.01333 | 4958 | |
| GC-skew reverse | 5358 | 0.00667 | 2859 |
| 6050 | 0 | 4707 | |
| AT-skew forward | 306 | 0 | 681 |
| 754 | 0 | 1641 | |
| 1146 | 0 | 2335 | |
| 2531 | 0 | 5076 | |
| AT-skew reverse | 4802 | 0 | 1407 |
| 5330 | 0.04889 | 2795 | |
| 6266 | 0.00222 | 5211 |
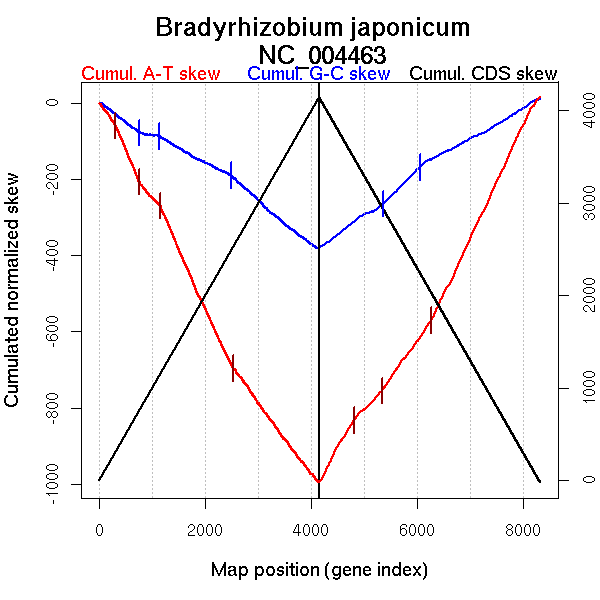
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 757 (1645.778 kb) | NA | -0.107 |
| 758 (1648.5805 kb) | 1135 (2319.173 kb) | leading | -0.02 | |
| 1136 (2320.658 kb) | 2482 (4957.524 kb) | leading | -0.076 | |
| 2483 (4958.895 kb) | 4147 (9105.797 kb) | lagging | -0.118 | |
| GC-skew reverse | 4148 (0 kb) | 5358 (2859.041 kb) | NA | 0.097 |
| 5359 (2859.841 kb) | 6050 (4707.2085 kb) | leading | 0.141 | |
| 6051 (4710.0715 kb) | 8317 (9105.797 kb) | lagging | 0.078 | |
| AT-skew forward | 1 (0 kb) | 306 (680.5335 kb) | lagging | -0.189 |
| 307 (685.99 kb) | 754 (1641.0115 kb) | leading | -0.331 | |
| 755 (1641.4245 kb) | 1146 (2334.5915 kb) | leading | -0.153 | |
| 1147 (2337.9065 kb) | 2531 (5076.3265 kb) | leading | -0.311 | |
| 2532 (5079.429 kb) | 4147 (9105.797 kb) | lagging | -0.189 | |
| AT-skew reverse | 4148 (0 kb) | 4802 (1407.391 kb) | NA | 0.255 |
| 4803 (1408.4165 kb) | 5330 (2794.706 kb) | leading | 0.138 | |
| 5331 (2796.3425 kb) | 6266 (5211.3115 kb) | leading | 0.194 | |
| 6267 (5212.6245 kb) | 8317 (9105.797 kb) | lagging | 0.295 |
More G than C on the leading strand for replication - for forward encoded genes.