Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Vibrio.
Genome size (bp): 1877212
Number of genes 1752
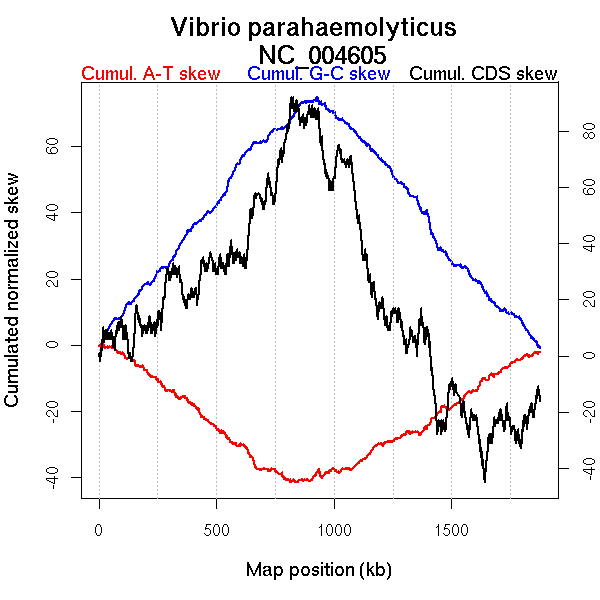
Oriloc predictions: Origin 0 kb Terminus 895 kb
Worning et al., 2006: Origin 0 kb Terminus 935 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 916.772 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 944.02 kb
Consensus predictions: Origin 0 kb Terminus 895 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 470 | NA | 890 |
| GC-skew reverse | 1277 | NA | 939 |
| AT-skew forward | 365 | NA | 699 |
| AT-skew reverse | 1242 | NA | 871 |
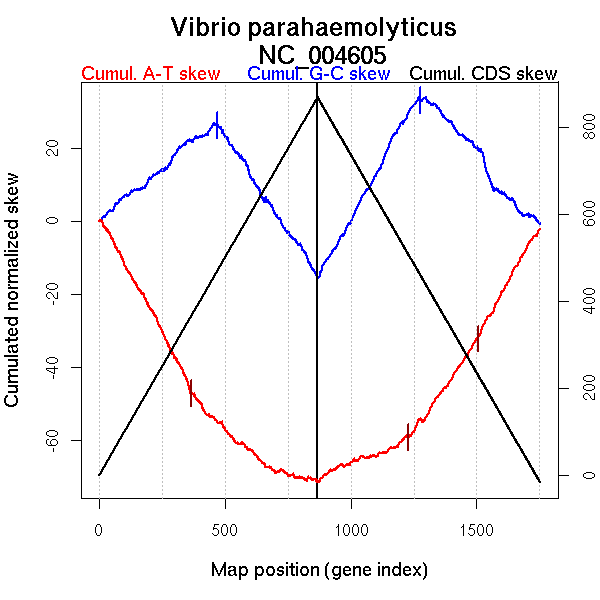
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 470 (890.2455 kb) | leading | 0.057 |
| 471 (893.33 kb) | 868 (1877.11 kb) | lagging | -0.104 | |
| GC-skew reverse | 869 (0 kb) | 1277 (939.4735 kb) | leading | 0.125 |
| 1278 (940.0345 kb) | 1752 (1877.11 kb) | lagging | -0.082 | |
| AT-skew forward | 1 (0 kb) | 365 (699.1065 kb) | leading | -0.128 |
| 366 (702.4925 kb) | 868 (1877.11 kb) | NA | -0.049 | |
| AT-skew reverse | 869 (0 kb) | 1242 (870.571 kb) | leading | 0.029 |
| 1243 (872.1275 kb) | 1752 (1877.11 kb) | lagging | 0.113 |
More G than C on the leading strand for replication.