Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Vibrio.
Genome size (bp): 3288558
Number of genes 3080
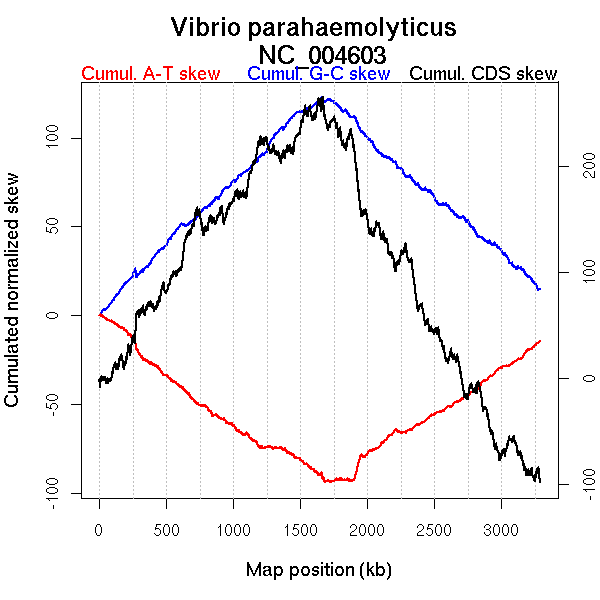
Oriloc predictions: Origin 0 kb Terminus 1707 kb
Worning et al., 2006: Origin 3287 kb Terminus 1667 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 0.219 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 8.38 kb, 1678.39 kb
Consensus predictions: Origin 0 kb Terminus 1707 kb
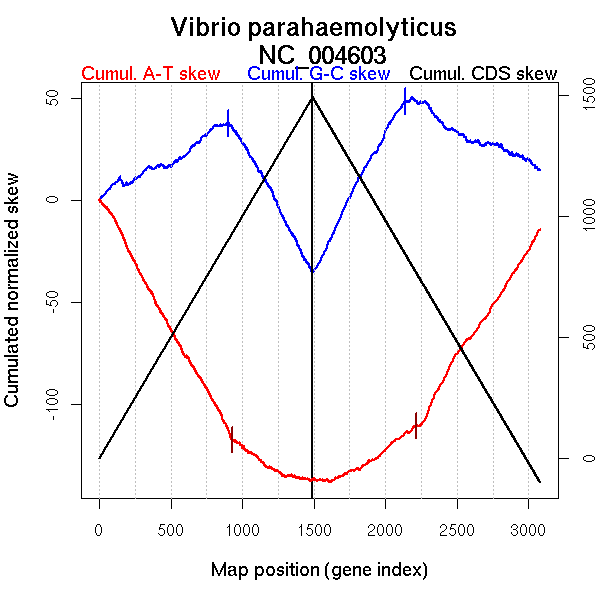
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 905 | NA | 1661 |
| GC-skew reverse | 2135 | NA | 1653 |
| AT-skew forward | 105 | NA | 202 |
| 290 | NA | 512 | |
| 927 | NA | 1717 | |
| 1305 | NA | 2762 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 905 (1660.9875 kb) | leading | 0.04 |
| 906 (1661.2225 kb) | 1491 (3288.454 kb) | lagging | -0.129 | |
| GC-skew reverse | 1492 (0 kb) | 2135 (1653.116 kb) | leading | 0.134 |
| 2136 (1653.9305 kb) | 3080 (3288.454 kb) | lagging | -0.036 | |
| AT-skew forward | 1 (0 kb) | 105 (202.292 kb) | leading | -0.081 |
| 106 (204.62 kb) | 290 (511.966 kb) | leading | -0.152 | |
| 291 (513.6665 kb) | 927 (1717.393 kb) | leading | -0.122 | |
| 928 (1718.648 kb) | 1305 (2761.9375 kb) | lagging | -0.05 | |
| 1306 (2762.4915 kb) | 1491 (3288.454 kb) | lagging | -0.014 |
More G than C on the leading strand for replication.