Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Vibrio.
Genome size (bp): 3354505
Number of genes 3259
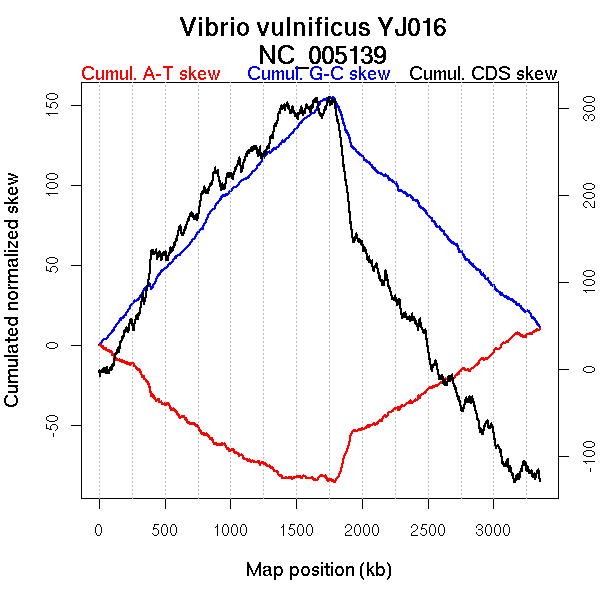
Oriloc predictions: Origin 0 kb Terminus 1777 kb
Worning et al., 2006: Origin 3353 kb Terminus 1743 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 0.211 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 8.15 kb
Consensus predictions: Origin 0 kb Terminus 1777 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 1006 | NA | 1762 |
| GC-skew reverse | 2264 | NA | 1757 |
| AT-skew forward | 177 | NA | 302 |
| 304 | NA | 460 | |
| 668 | NA | 1102 | |
| 1022 | NA | 1791 |
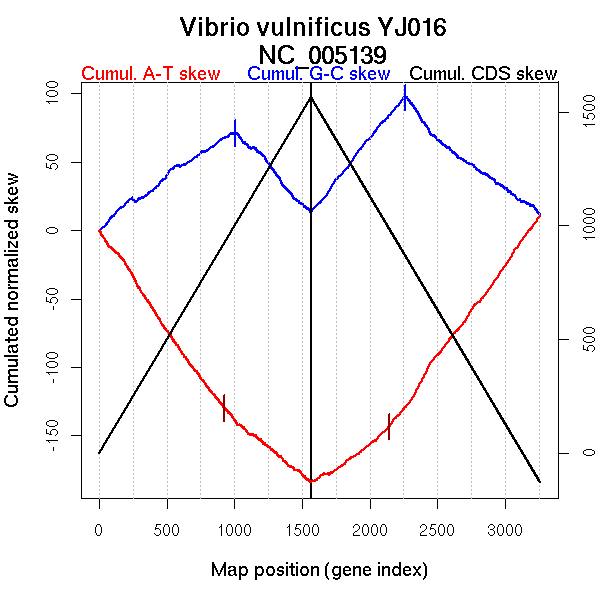
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 1006 (1761.8195 kb) | leading | 0.07 |
| 1007 (1765.1285 kb) | 1565 (3354.389 kb) | lagging | -0.11 | |
| GC-skew reverse | 1566 (0 kb) | 2264 (1757.347 kb) | leading | 0.123 |
| 2265 (1759.984 kb) | 3259 (3354.389 kb) | lagging | -0.079 | |
| AT-skew forward | 1 (0 kb) | 177 (302.217 kb) | leading | -0.114 |
| 178 (304.499 kb) | 304 (459.7335 kb) | leading | -0.176 | |
| 305 (463.7275 kb) | 668 (1101.7975 kb) | leading | -0.148 | |
| 669 (1103.258 kb) | 1022 (1791.485 kb) | leading | -0.124 | |
| 1023 (1796.212 kb) | 1565 (3354.389 kb) | lagging | -0.079 |
More G than C on the leading strand for replication.