Rearranged Oriloc output example: Previous Index Next
Bacteria; Actinobacteria; Actinobacteridae; Actinomycetales; Micrococcineae; Microbacteriaceae; Leifsonia.
Genome size (bp): 2584158
Number of genes 2030
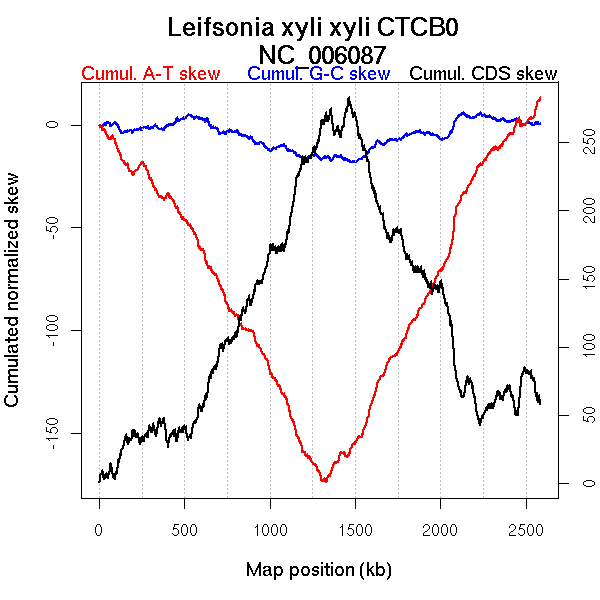
Oriloc predictions: Origin 30 kb Terminus 1301 kb
Worning et al., 2006: Origin 2573 kb Terminus 1329 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 17.296 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.94 kb
Consensus predictions: Origin 30 kb Terminus 1301 kb
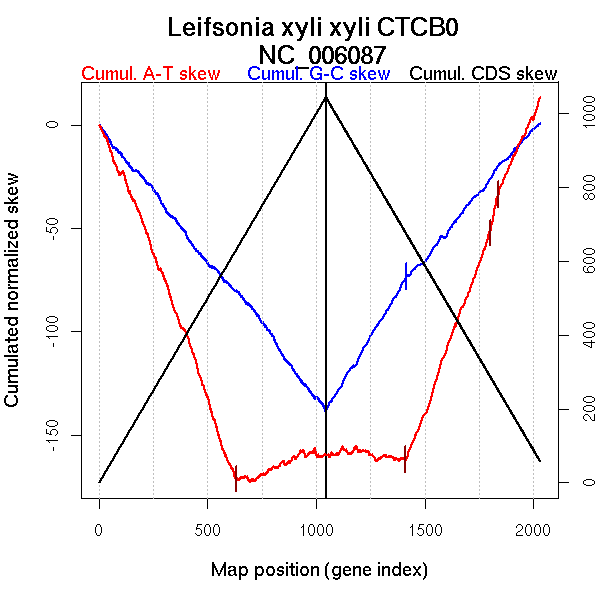
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew reverse | 1416 | 0.01333 | 1334 |
| AT-skew forward | 629 | 0 | 1312 |
| AT-skew reverse | 1408 | 0 | 1311 |
| 1799 | 0.00889 | 2072 | |
| 1838 | 0 | 2108 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew reverse | 1045 (0 kb) | 1416 (1333.8965 kb) | leading | 0.173 |
| 1417 (1335.0235 kb) | 2030 (2583.936 kb) | lagging | 0.126 | |
| AT-skew forward | 1 (0 kb) | 629 (1312.3865 kb) | leading | -0.277 |
| 630 (1316.2955 kb) | 1044 (2583.936 kb) | lagging | 0.039 | |
| AT-skew reverse | 1045 (0 kb) | 1408 (1310.9005 kb) | leading | -0.01 |
| 1409 (1313.2485 kb) | 1799 (2071.5535 kb) | lagging | 0.286 | |
| 1800 (2072.037 kb) | 1838 (2108.4795 kb) | lagging | 0.516 | |
| 1839 (2112.21 kb) | 2030 (2583.936 kb) | lagging | 0.228 |
More G than C on the leading strand for replication - for reverse encoded genes.