Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Gluconobacter.
Genome size (bp): 2702173
Number of genes 2432
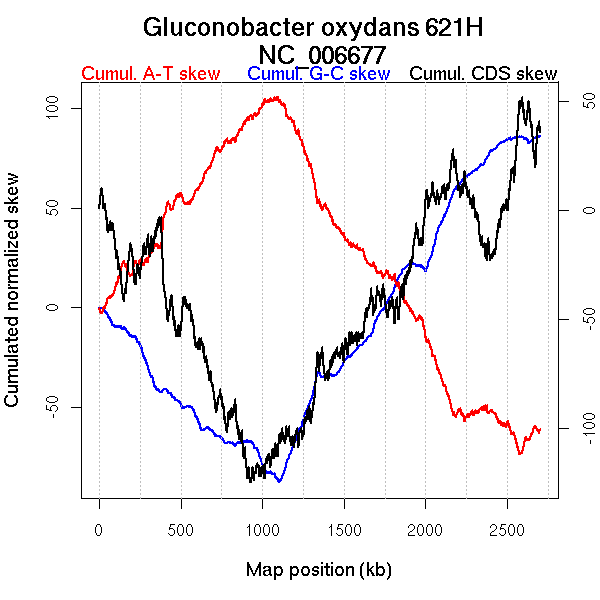
Oriloc predictions: Origin 1101 kb Terminus 2586 kb
Worning et al., 2006: Origin 1102 kb Terminus 2571 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 447.934 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 2124.48 kb
Consensus predictions: Origin 1101 kb Terminus 2586 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 435 | 0 | 1101 |
| 561 | 0.02 | 1338 | |
| 1101 | 0.03556 | 2486 | |
| GC-skew reverse | 1783 | 0 | 1124 |
| 2350 | 0 | 2536 | |
| AT-skew forward | 443 | 0.00667 | 1116 |
| 1039 | 0.00667 | 2371 | |
| AT-skew reverse | 1782 | 0.00667 | 1117 |
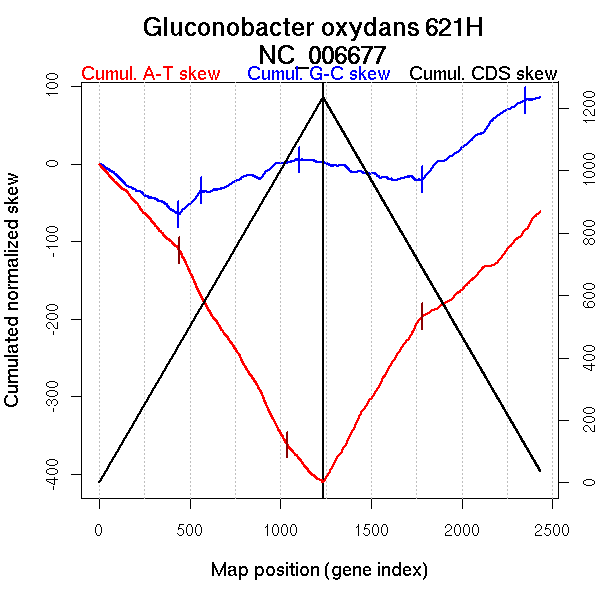
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 435 (1100.6995 kb) | lagging | -0.144 |
| 436 (1101.538 kb) | 561 (1337.572 kb) | leading | 0.237 | |
| 562 (1338.2605 kb) | 1101 (2486.177 kb) | leading | 0.086 | |
| 1102 (2488.612 kb) | 1235 (2702 kb) | NA | -0.031 | |
| GC-skew reverse | 1236 (0 kb) | 1783 (1123.75 kb) | lagging | -0.042 |
| 1784 (1125.099 kb) | 2350 (2536.484 kb) | leading | 0.181 | |
| 2351 (2539.1845 kb) | 2432 (2702 kb) | lagging | 0.044 | |
| AT-skew forward | 1 (0 kb) | 443 (1116.246 kb) | lagging | -0.253 |
| 444 (1117.5095 kb) | 1039 (2371.359 kb) | leading | -0.404 | |
| 1040 (2373.823 kb) | 1235 (2702 kb) | NA | -0.253 | |
| AT-skew reverse | 1236 (0 kb) | 1782 (1116.5155 kb) | lagging | 0.395 |
| 1783 (1123.75 kb) | 2432 (2702 kb) | leading | 0.208 |
More G than C on the leading strand for replication.