Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Gammaproteobacteria; Pasteurellales; Pasteurellaceae; Haemophilus.
Genome size (bp): 1698955
Number of genes 1717
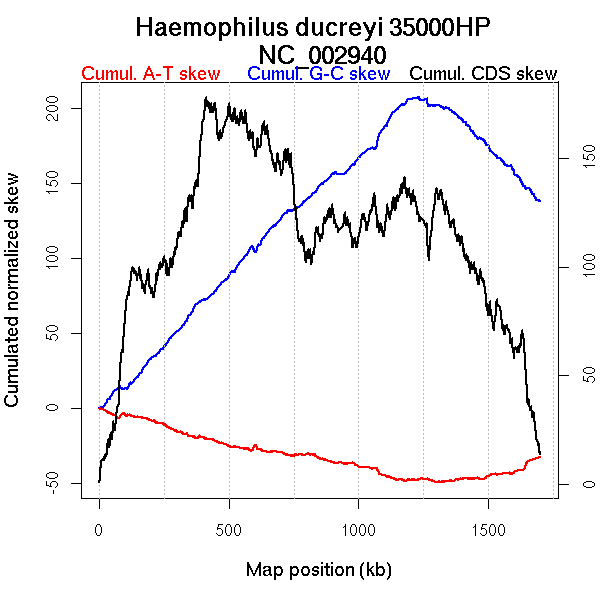
Oriloc predictions: Origin 0 kb Terminus 1216 kb
Worning et al., 2006: Origin 0 kb Terminus 1178 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 1698.947 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 679.1 kb
Consensus predictions: Origin 0 kb Terminus 1216 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 52 | 0.01167 | 71 |
| 108 | 0.03167 | 109 | |
| 682 | 0.035 | 1187 | |
| 766 | 0.04667 | 1395 | |
| GC-skew reverse | 1422 | 0 | 1201 |
| AT-skew forward | 650 | 0.01333 | 1129 |
| AT-skew reverse | 1482 | 0 | 1328 |
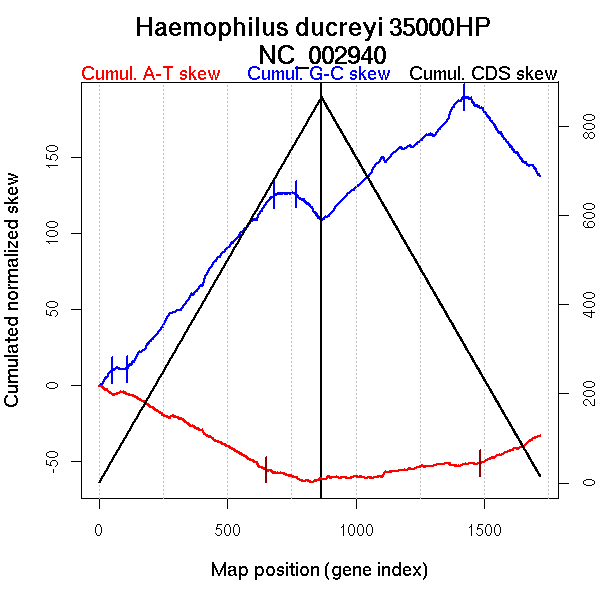
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 52 (70.889 kb) | leading | 0.225 |
| 53 (72.1755 kb) | 108 (108.766 kb) | leading | 0.002 | |
| 109 (109.6105 kb) | 682 (1186.7265 kb) | leading | 0.2 | |
| 683 (1188.2945 kb) | 766 (1395.4795 kb) | lagging | 0.01 | |
| 767 (1400.4895 kb) | 866 (1698.704 kb) | lagging | -0.17 | |
| GC-skew reverse | 867 (0 kb) | 1422 (1201.151 kb) | leading | 0.134 |
| 1423 (1202.06 kb) | 1717 (1698.704 kb) | lagging | -0.191 | |
| AT-skew forward | 1 (0 kb) | 650 (1129.358 kb) | leading | -0.085 |
| 651 (1130.748 kb) | 866 (1698.704 kb) | NA | -0.037 | |
| AT-skew reverse | 867 (0 kb) | 1482 (1327.985 kb) | leading | 0.02 |
| 1483 (1329.267 kb) | 1717 (1698.704 kb) | lagging | 0.075 |
More G than C on the leading strand for replication.