Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae; ant endosymbionts; Candidatus Blochmannia.
Genome size (bp): 791654
Number of genes 606
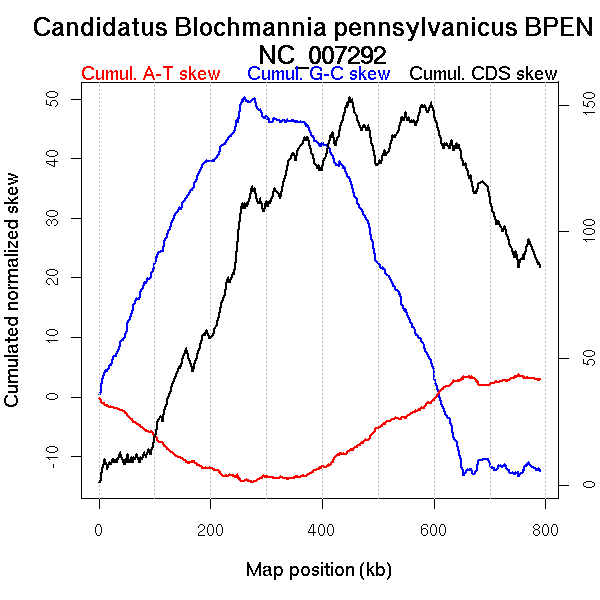
Oriloc predictions: Origin 0 kb Terminus 270 kb
Worning et al., 2006: Origin 749 kb Terminus 280 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 330.744 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): NA
Consensus predictions: Origin 0 kb Terminus 270 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 97 | NA | 182 |
| 210 | NA | 359 | |
| 319 | NA | 670 | |
| GC-skew reverse | 410 | NA | 292 |
| 537 | NA | 655 | |
| AT-skew forward | 101 | NA | 190 |
| 208 | NA | 355 | |
| AT-skew reverse | 397 | NA | 262 |
| 531 | NA | 648 |
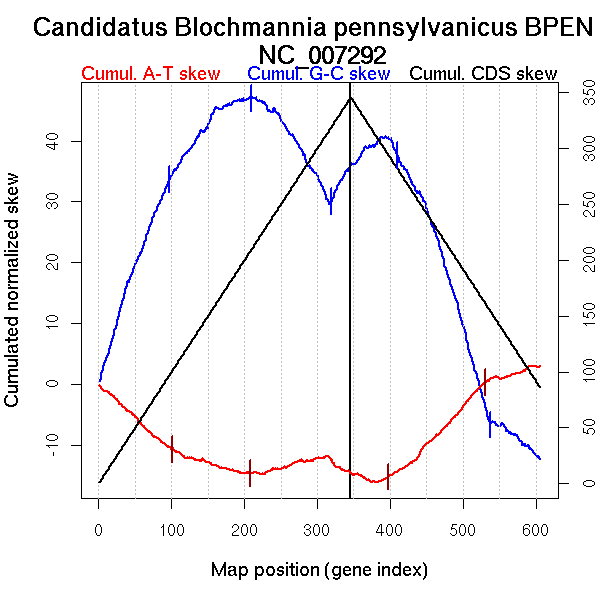
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 97 (182.487 kb) | leading | 0.345 |
| 98 (183.7985 kb) | 210 (359.223 kb) | NA | 0.123 | |
| 211 (363.0835 kb) | 319 (669.8115 kb) | lagging | -0.169 | |
| 320 (675.0555 kb) | 346 (790.625 kb) | lagging | 0.211 | |
| GC-skew reverse | 347 (0 kb) | 410 (292.448 kb) | NA | 0.054 |
| 411 (293.64 kb) | 537 (655.146 kb) | lagging | -0.358 | |
| 538 (657.8235 kb) | 606 (790.625 kb) | lagging | -0.093 | |
| AT-skew forward | 1 (0 kb) | 101 (190.285 kb) | leading | -0.109 |
| 102 (191.613 kb) | 208 (355.1035 kb) | NA | -0.036 | |
| 209 (356.9935 kb) | 346 (790.625 kb) | lagging | 0.011 | |
| AT-skew reverse | 347 (0 kb) | 397 (261.687 kb) | leading | -0.022 |
| 398 (263.5255 kb) | 531 (647.5475 kb) | lagging | 0.123 | |
| 532 (648.2815 kb) | 606 (790.625 kb) | lagging | 0.035 |
More G than C on the leading strand for replication - for forward encoded genes.