Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae; ant endosymbionts; Candidatus Blochmannia.
Genome size (bp): 705557
Number of genes 583
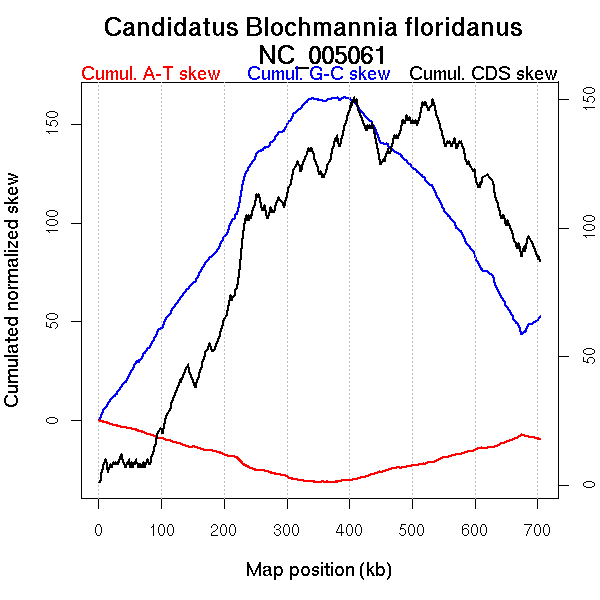
Oriloc predictions: Origin 675 kb Terminus 376 kb
Worning et al., 2006: Origin 675 kb Terminus 372 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 255.113 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): NA
Consensus predictions: Origin 675 kb Terminus 376 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 199 | 0.02 | 324 |
| 245 | 0.04444 | 403 | |
| 327 | 0 | 678 | |
| GC-skew reverse | 408 | 0.03 | 320 |
| 439 | 0.03667 | 415 | |
| AT-skew forward | 197 | 0.04 | 316 |
| 226 | 0.04667 | 378 | |
| AT-skew reverse | 422 | 0.02333 | 351 |
| 573 | 0.01 | 688 |
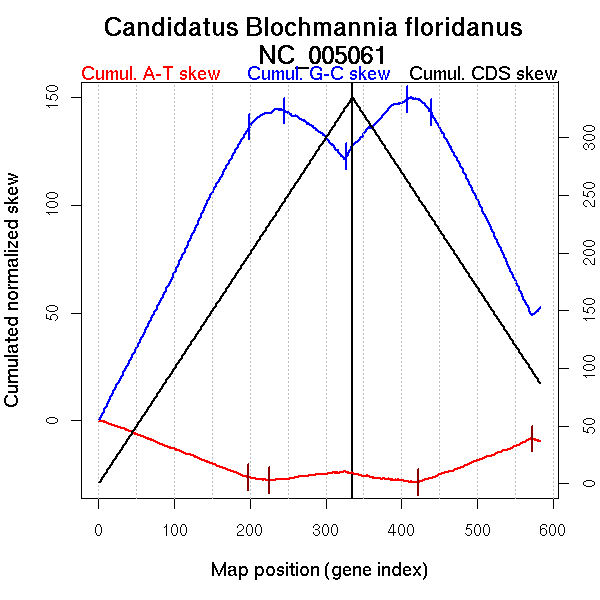
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 199 (323.94 kb) | leading | 0.701 |
| 200 (324.8235 kb) | 245 (403.289 kb) | NA | 0.147 | |
| 246 (404.311 kb) | 327 (678.025 kb) | lagging | -0.279 | |
| 328 (679.958 kb) | 335 (704.748 kb) | leading | 0.655 | |
| GC-skew reverse | 336 (0 kb) | 408 (319.584 kb) | leading | 0.307 |
| 409 (321.129 kb) | 439 (415.1275 kb) | NA | -0.223 | |
| 440 (416.7295 kb) | 583 (704.748 kb) | lagging | -0.703 | |
| AT-skew forward | 1 (0 kb) | 197 (315.7665 kb) | leading | -0.137 |
| 198 (320.158 kb) | 226 (378.097 kb) | leading | -0.045 | |
| 227 (379.5185 kb) | 335 (704.748 kb) | lagging | 0.039 | |
| AT-skew reverse | 336 (0 kb) | 422 (351.387 kb) | leading | -0.043 |
| 423 (353.3365 kb) | 573 (688.219 kb) | lagging | 0.134 | |
| 574 (690.9225 kb) | 583 (704.748 kb) | leading | -0.107 |
More G than C on the leading strand for replication.