Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Betaproteobacteria; Rhodocyclales; Rhodocyclaceae; Dechloromonas.
Genome size (bp): 4501104
Number of genes 4171
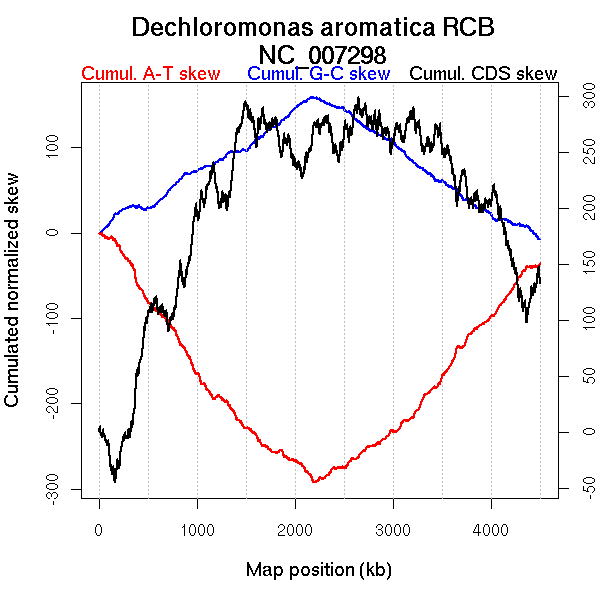
Oriloc predictions: Origin 4 kb Terminus 2193 kb
Worning et al., 2006: Origin 4501 kb Terminus 2194 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 2687.74 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.85 kb
Consensus predictions: Origin 4 kb Terminus 2193 kb
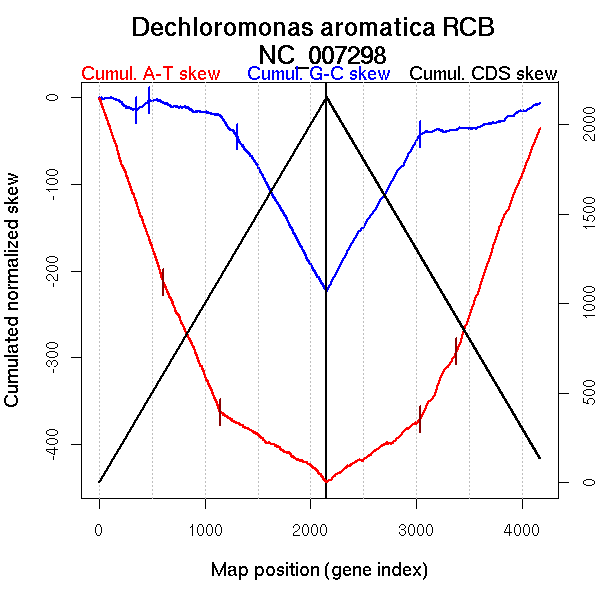
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 354 | 0 | 675 |
| 473 | 0 | 882 | |
| 1307 | 0 | 2557 | |
| GC-skew reverse | 3040 | 0 | 2223 |
| AT-skew forward | 609 | 0.04667 | 1097 |
| 1147 | 0.00667 | 2187 | |
| AT-skew reverse | 3035 | 0 | 2215 |
| 3382 | 0.02333 | 2957 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 354 (675.027 kb) | leading | -0.048 |
| 355 (678.2485 kb) | 473 (881.921 kb) | leading | 0.093 | |
| 474 (882.4175 kb) | 1307 (2557.494 kb) | NA | -0.04 | |
| 1308 (2557.76 kb) | 2152 (4501.019 kb) | lagging | -0.217 | |
| GC-skew reverse | 2153 (0 kb) | 3040 (2223.1145 kb) | leading | 0.198 |
| 3041 (2227.331 kb) | 4171 (4501.019 kb) | lagging | 0.029 | |
| AT-skew forward | 1 (0 kb) | 609 (1097.21 kb) | leading | -0.346 |
| 610 (1097.9375 kb) | 1147 (2186.6865 kb) | leading | -0.275 | |
| 1148 (2188.2145 kb) | 2152 (4501.019 kb) | lagging | -0.074 | |
| AT-skew reverse | 2153 (0 kb) | 3035 (2215.2505 kb) | leading | 0.078 |
| 3036 (2216.2995 kb) | 3382 (2957.422 kb) | lagging | 0.229 | |
| 3383 (2958.58 kb) | 4171 (4501.019 kb) | lagging | 0.327 |
More G than C on the leading strand for replication - for reverse encoded genes.