Rearranged Oriloc output example: Previous Index Next
Bacteria; Actinobacteria; Actinobacteridae; Actinomycetales; Streptosporangineae; Nocardiopsaceae; Thermobifida.
Genome size (bp): 3642249
Number of genes 3110
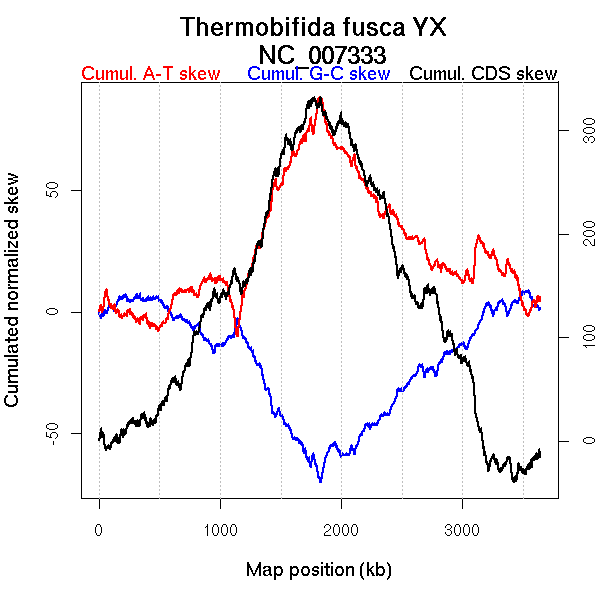
Oriloc predictions: Origin 3497 kb Terminus 1828 kb
Worning et al., 2006: Origin 1829 kb Terminus 3507 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 2.227 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 1.12 kb
Consensus predictions: Origin 3497 kb Terminus 1828 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew reverse | 1905 | 0.025 | 1012 |
| 1948 | 0.01 | 1133 | |
| 2182 | 0.00167 | 1841 | |
| 2209 | 0.00667 | 1903 | |
| AT-skew forward | 917 | 0.00667 | 1744 |
| AT-skew reverse | 1946 | 0.04667 | 1128 |
| 2186 | 0.04333 | 1847 |
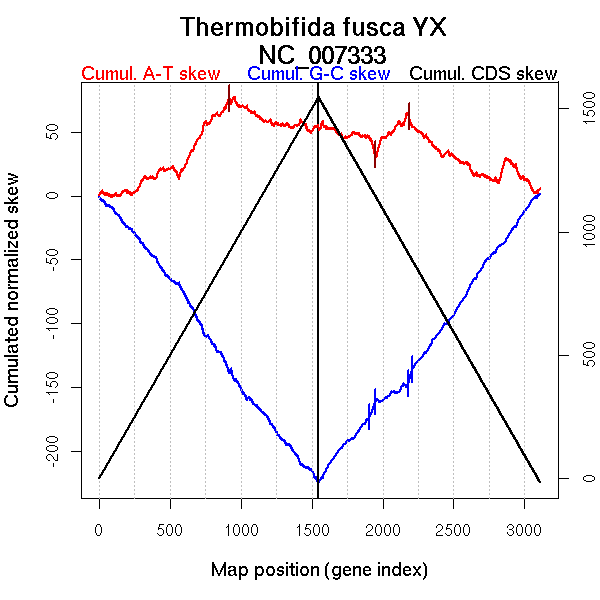
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew reverse | 1548 (0 kb) | 1905 (1012.204 kb) | leading | 0.135 |
| 1906 (1014.24 kb) | 1948 (1132.7905 kb) | leading | 0.276 | |
| 1949 (1133.404 kb) | 2182 (1841.465 kb) | leading | 0.063 | |
| 2183 (1842.1525 kb) | 2209 (1902.707 kb) | lagging | 0.416 | |
| 2210 (1905.343 kb) | 3110 (3641.655 kb) | lagging | 0.157 | |
| AT-skew forward | 1 (0 kb) | 917 (1744.0325 kb) | leading | 0.081 |
| 918 (1747.9295 kb) | 1547 (3641.655 kb) | NA | -0.035 | |
| AT-skew reverse | 1548 (0 kb) | 1946 (1128.164 kb) | leading | -0.031 |
| 1947 (1130.325 kb) | 2186 (1847.268 kb) | leading | 0.118 | |
| 2187 (1850.481 kb) | 3110 (3641.655 kb) | lagging | -0.047 |
More C than G on the leading strand for replication - for reverse encoded genes.