Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Bradyrhizobiaceae; Nitrobacter.
Genome size (bp): 3402093
Number of genes 3122
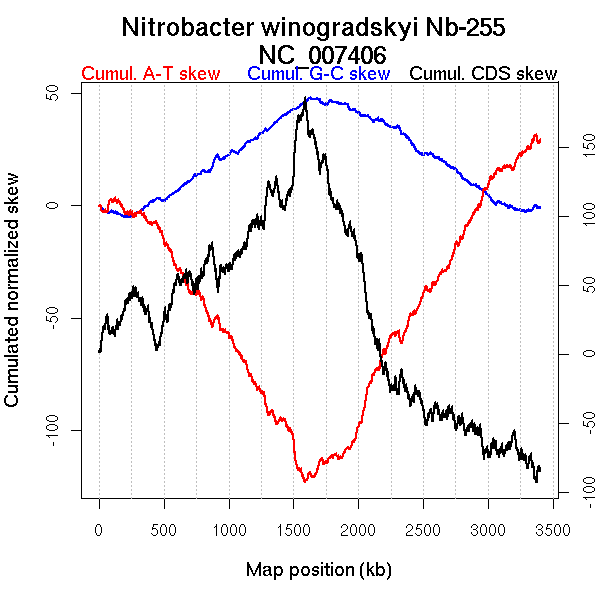
Oriloc predictions: Origin 0 kb Terminus 1613 kb
Worning et al., 2006: Origin 241 kb Terminus 1591 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 2366.634 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 1.28 kb, 2420.74 kb
Consensus predictions: Origin 0 kb Terminus 1613 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 824 | NA | 1628 |
| GC-skew reverse | 2168 | NA | 1608 |
| AT-skew forward | 920 | NA | 1926 |
| AT-skew reverse | 2334 | NA | 1892 |
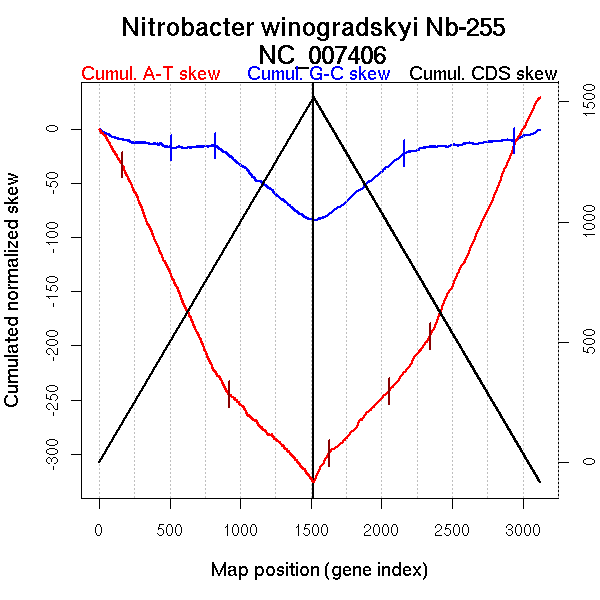
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 824 (1627.8905 kb) | leading | -0.016 |
| 825 (1628.5305 kb) | 1519 (3401.218 kb) | lagging | -0.105 | |
| GC-skew reverse | 1520 (0 kb) | 2168 (1608.32 kb) | leading | 0.103 |
| 2169 (1608.6865 kb) | 3122 (3401.218 kb) | lagging | 0.016 | |
| AT-skew forward | 1 (0 kb) | 920 (1925.655 kb) | leading | -0.282 |
| 921 (1926.838 kb) | 1519 (3401.218 kb) | lagging | -0.131 | |
| AT-skew reverse | 1520 (0 kb) | 2334 (1892.367 kb) | NA | 0.151 |
| 2335 (1893.438 kb) | 3122 (3401.218 kb) | lagging | 0.288 |
More G than C on the leading strand for replication.