Rearranged Oriloc output example: Previous Index Next
Bacteria; Cyanobacteria; Chroococcales; Synechococcus.
Genome size (bp): 2510659
Number of genes 2645
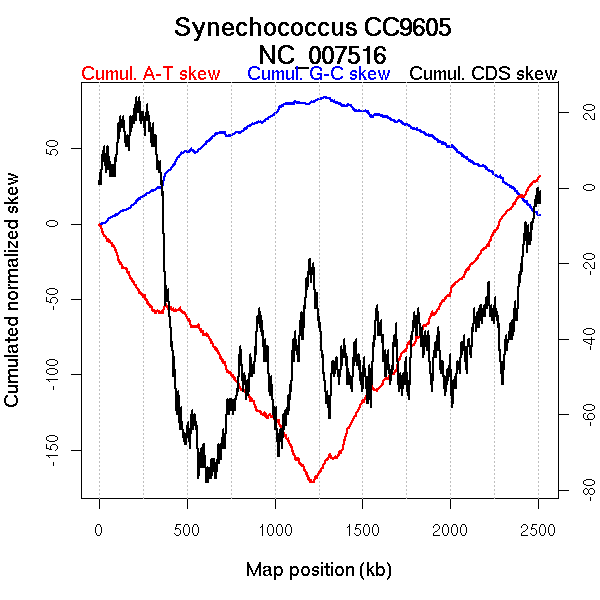
Oriloc predictions: Origin 0 kb Terminus 1203 kb
Worning et al., 2006: Origin 1 kb Terminus 1240 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 517.486 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 928.44 kb
Consensus predictions: Origin 0 kb Terminus 1203 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 521 | 0.02 | 1059 |
| 988 | 0.03333 | 1873 | |
| GC-skew reverse | 1587 | 0.04667 | 464 |
| 2003 | 0.03 | 1262 | |
| AT-skew forward | 629 | 0 | 1220 |
| AT-skew reverse | 1973 | 0 | 1222 |
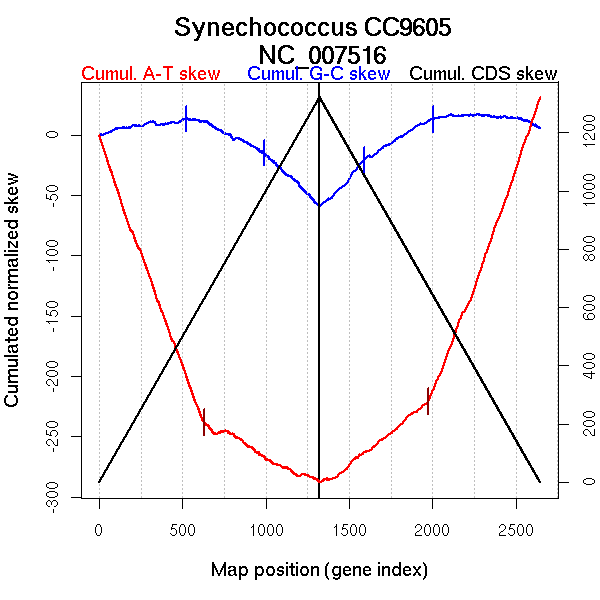
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 521 (1058.612 kb) | leading | 0.02 |
| 522 (1062.5965 kb) | 988 (1873.152 kb) | NA | -0.064 | |
| 989 (1875.07 kb) | 1322 (2510.454 kb) | lagging | -0.13 | |
| GC-skew reverse | 1323 (0 kb) | 1587 (463.7185 kb) | leading | 0.146 |
| 1588 (466.006 kb) | 2003 (1261.969 kb) | leading | 0.085 | |
| 2004 (1262.572 kb) | 2645 (2510.454 kb) | lagging | -0.009 | |
| AT-skew forward | 1 (0 kb) | 629 (1219.9745 kb) | leading | -0.376 |
| 630 (1226.698 kb) | 1322 (2510.454 kb) | lagging | -0.073 | |
| AT-skew reverse | 1323 (0 kb) | 1973 (1221.826 kb) | leading | 0.108 |
| 1974 (1223.04 kb) | 2645 (2510.454 kb) | lagging | 0.376 |
More G than C on the leading strand for replication - for reverse encoded genes.