Rearranged Oriloc output example: Previous Index Next
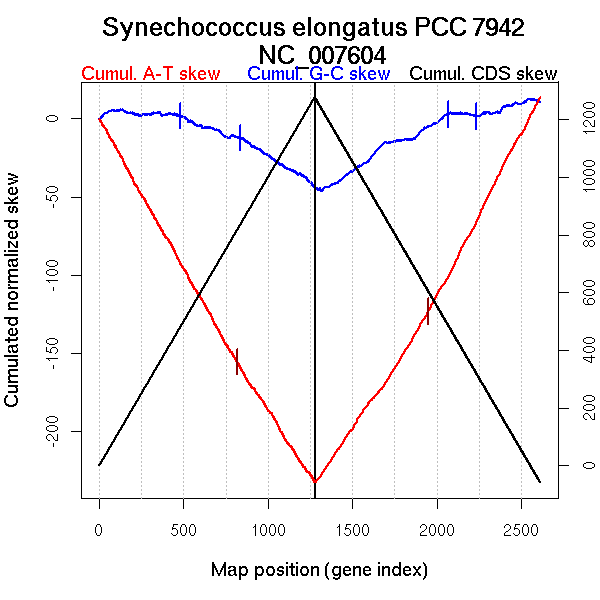
Bacteria; Cyanobacteria; Chroococcales; Synechococcus.
Genome size (bp): 2695903
Number of genes 2612
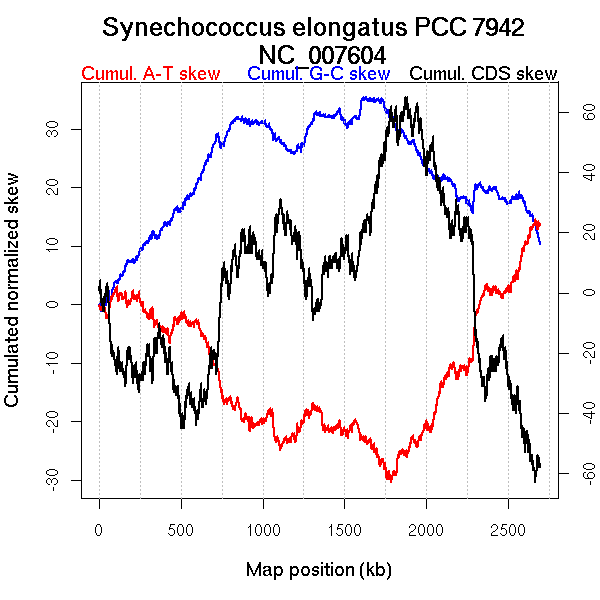
Oriloc predictions: Origin 24 kb Terminus 1715 kb
Worning et al., 2006: Origin 25 kb Terminus 1783 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 2695.801 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 1117.83 kb
Consensus predictions: Origin 0 kb Terminus 1715 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 481 | NA | 958 |
| GC-skew reverse | 2081 | NA | 1725 |
| AT-skew forward | 819 | NA | 1680 |
| AT-skew reverse | 1851 | NA | 1203 |
| 2074 | NA | 1703 | |
| 2435 | NA | 2357 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 481 (957.731 kb) | leading | -0.003 |
| 482 (962.304 kb) | 1277 (2695.87 kb) | NA | -0.054 | |
| GC-skew reverse | 1278 (0 kb) | 2081 (1725.3405 kb) | leading | 0.063 |
| 2082 (1726.672 kb) | 2612 (2695.87 kb) | lagging | 0.021 | |
| AT-skew forward | 1 (0 kb) | 819 (1680.454 kb) | leading | -0.191 |
| 820 (1682.6395 kb) | 1277 (2695.87 kb) | lagging | -0.167 | |
| AT-skew reverse | 1278 (0 kb) | 1851 (1203.212 kb) | leading | 0.157 |
| 1852 (1204.9745 kb) | 2074 (1703.2925 kb) | leading | 0.19 | |
| 2075 (1703.9195 kb) | 2435 (2357.0485 kb) | lagging | 0.218 | |
| 2436 (2357.9235 kb) | 2612 (2695.87 kb) | lagging | 0.198 |
More G than C on the leading strand for replication - for reverse encoded genes.