Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae; Magnetospirillum.
Genome size (bp): 4967148
Number of genes 4559
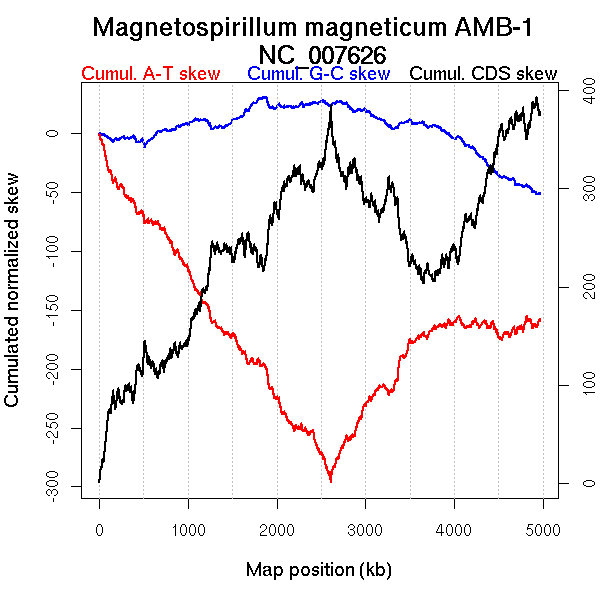
Oriloc predictions: Origin 0 kb Terminus 2611 kb
Worning et al., 2006: Origin 157 kb Terminus 2611 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 4958.066 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 3044.8 kb
Consensus predictions: Origin 0 kb Terminus 2611 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 1317 | 0 | 2513 |
| GC-skew reverse | 3486 | 0 | 2611 |
| AT-skew forward | 1420 | 0.01333 | 2690 |
| 2171 | 0.03333 | 4434 | |
| AT-skew reverse | 3464 | 0 | 2522 |
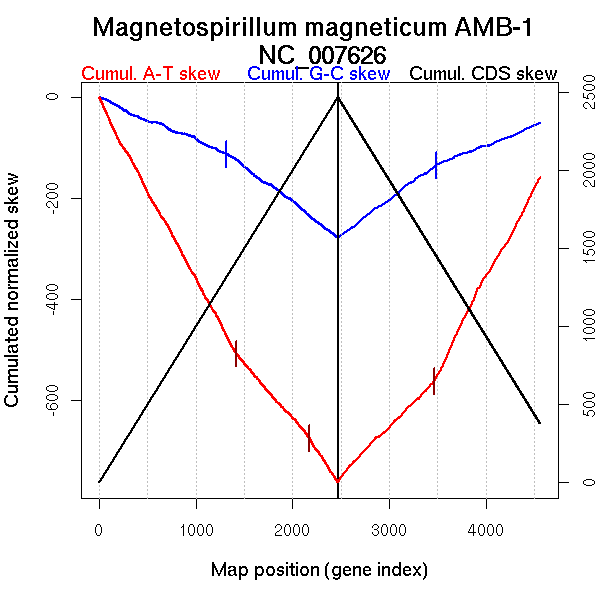
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 1317 (2512.911 kb) | leading | -0.083 |
| 1318 (2514.1815 kb) | 2468 (4967.145 kb) | lagging | -0.145 | |
| GC-skew reverse | 2469 (0 kb) | 3486 (2610.5265 kb) | leading | 0.142 |
| 3487 (2611.3545 kb) | 4559 (4967.145 kb) | lagging | 0.075 | |
| AT-skew forward | 1 (0 kb) | 1420 (2690.2615 kb) | leading | -0.352 |
| 1421 (2691.626 kb) | 2171 (4433.581 kb) | lagging | -0.217 | |
| 2172 (4434.667 kb) | 2468 (4967.145 kb) | lagging | -0.293 | |
| AT-skew reverse | 2469 (0 kb) | 3464 (2522.4585 kb) | leading | 0.192 |
| 3465 (2528.039 kb) | 4559 (4967.145 kb) | lagging | 0.362 |
More G than C on the leading strand for replication.