Rearranged Oriloc output example: Previous Index Next
Bacteria; Firmicutes; Clostridia; Thermoanaerobacteriales; Thermoanaerobacteriaceae; Moorella group; Moorella.
Genome size (bp): 2628784
Number of genes 2465
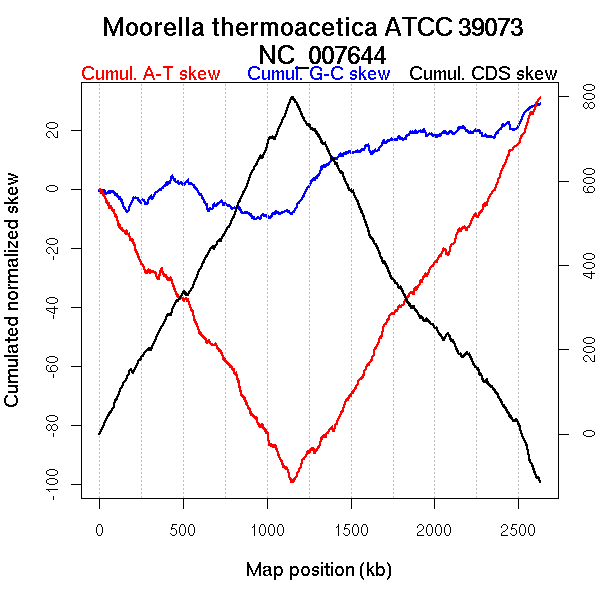
Oriloc predictions: Origin 0 kb Terminus 1147 kb
Worning et al., 2006: Origin 1 kb Terminus 1148 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 1166.999 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 1.11 kb
Consensus predictions: Origin 0 kb Terminus 1147 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 956 | 0 | 1245 |
| GC-skew reverse | 1506 | 0.01778 | 1388 |
| 1699 | 0 | 1606 | |
| 1716 | 0 | 1619 | |
| AT-skew forward | 233 | 0.00667 | 283 |
| 333 | 0.00667 | 401 | |
| AT-skew reverse | 1294 | 0 | 939 |
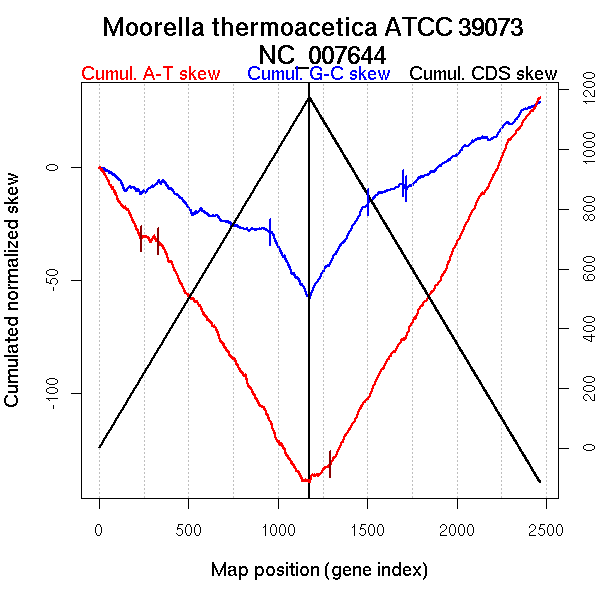
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 956 (1245.3885 kb) | leading | -0.031 |
| 957 (1246.805 kb) | 1175 (2628.756 kb) | lagging | -0.134 | |
| GC-skew reverse | 1176 (0 kb) | 1506 (1388.105 kb) | NA | 0.125 |
| 1507 (1388.6975 kb) | 1699 (1606.0695 kb) | lagging | 0.039 | |
| 1700 (1606.4245 kb) | 1716 (1618.614 kb) | lagging | -0.148 | |
| 1717 (1619.487 kb) | 2465 (2628.756 kb) | lagging | 0.05 | |
| AT-skew forward | 1 (0 kb) | 233 (283.4095 kb) | leading | -0.131 |
| 234 (283.7385 kb) | 333 (401.0435 kb) | leading | -0.015 | |
| 334 (402.0475 kb) | 1175 (2628.756 kb) | NA | -0.13 | |
| AT-skew reverse | 1176 (0 kb) | 1294 (938.6205 kb) | leading | 0.056 |
| 1295 (946.949 kb) | 2465 (2628.756 kb) | lagging | 0.14 |
More G than C on the leading strand for replication - for forward encoded genes.