Rearranged Oriloc output example: Previous Index Next
Bacteria; Actinobacteria; Actinobacteridae; Actinomycetales; Corynebacterineae; Mycobacteriaceae; Mycobacterium.
Genome size (bp): 5705448
Number of genes 5391
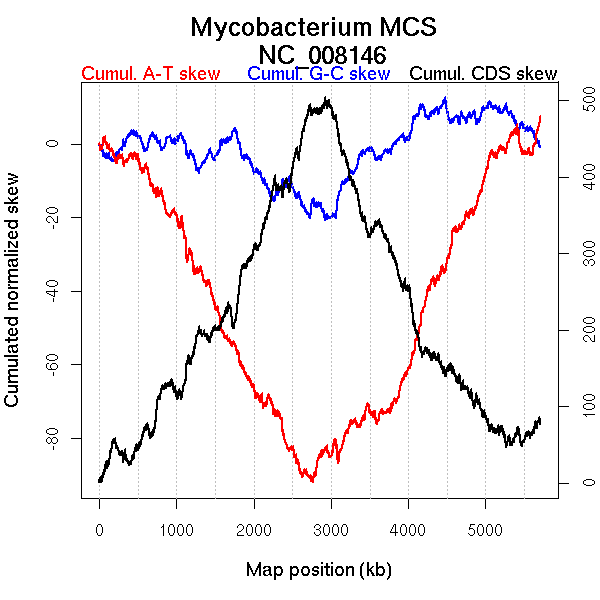
Oriloc predictions: Origin 0 kb Terminus 2672 kb
Worning et al., 2006: Origin NA kb Terminus NA kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 5654.273 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.76 kb
Consensus predictions: Origin 0 kb Terminus 2672 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 1452 | 0.00667 | 2622 |
| GC-skew reverse | 4174 | 0.00667 | 3437 |
| AT-skew forward | 1432 | 0.00667 | 2594 |
| AT-skew reverse | 3975 | 0 | 3106 |
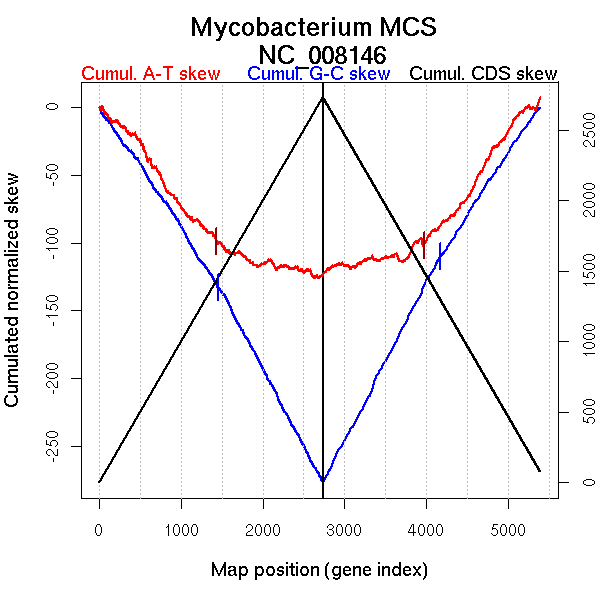
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 1452 (2621.8515 kb) | leading | -0.091 |
| 1453 (2622.7795 kb) | 2734 (5704.914 kb) | lagging | -0.114 | |
| GC-skew reverse | 2735 (0 kb) | 4174 (3436.8765 kb) | NA | 0.117 |
| 4175 (3439.643 kb) | 5391 (5704.914 kb) | lagging | 0.092 | |
| AT-skew forward | 1 (0 kb) | 1432 (2593.722 kb) | leading | -0.074 |
| 1433 (2595.4805 kb) | 2734 (5704.914 kb) | lagging | -0.017 | |
| AT-skew reverse | 2735 (0 kb) | 3975 (3106.034 kb) | NA | 0.014 |
| 3976 (3107.885 kb) | 5391 (5704.914 kb) | lagging | 0.081 |
More G than C on the leading strand for replication - for forward encoded genes.