Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae; Sodalis.
Genome size (bp): 4171146
Number of genes 2432
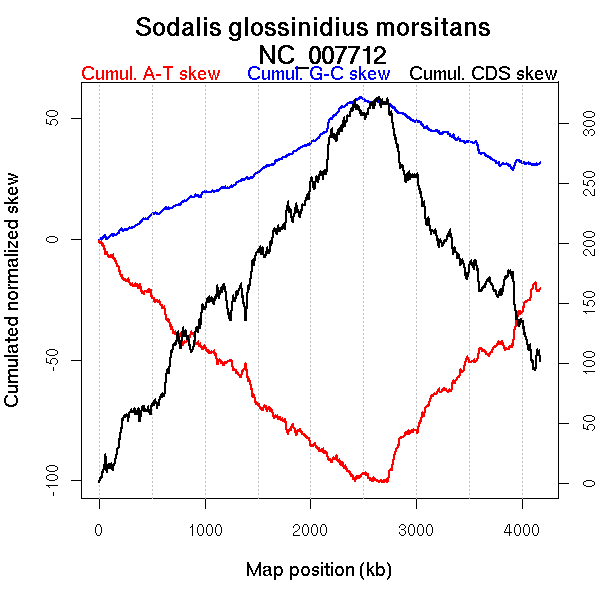
Oriloc predictions: Origin 0 kb Terminus 2461 kb
Worning et al., 2006: Origin 4128 kb Terminus 2463 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 4127.636 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.7 kb
Consensus predictions: Origin 0 kb Terminus 2461 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 725 | 0.02 | 2007 |
| 839 | 0 | 2294 | |
| GC-skew reverse | 1853 | 0 | 2475 |
| AT-skew forward | 684 | 0.01333 | 1904 |
| AT-skew reverse | 1898 | 0.00667 | 2710 |
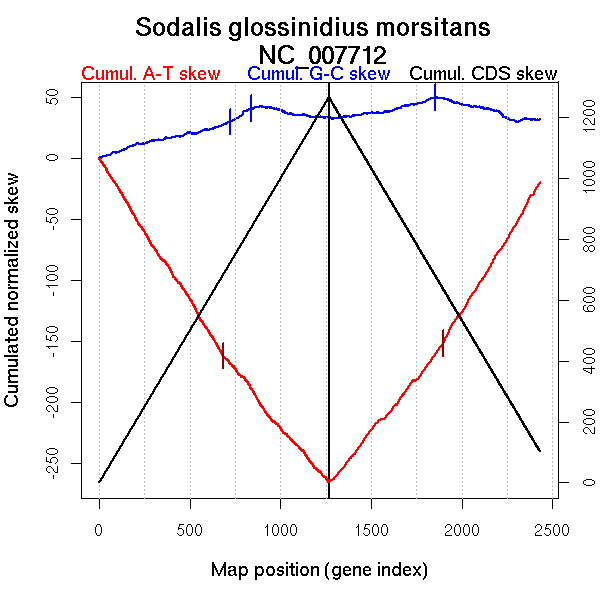
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 725 (2007.235 kb) | leading | 0.037 |
| 726 (2009.1665 kb) | 839 (2293.9915 kb) | leading | 0.104 | |
| 840 (2299.0505 kb) | 1267 (4170.51 kb) | lagging | -0.025 | |
| GC-skew reverse | 1268 (0 kb) | 1853 (2474.602 kb) | leading | 0.028 |
| 1854 (2476.211 kb) | 2432 (4170.51 kb) | lagging | -0.037 | |
| AT-skew forward | 1 (0 kb) | 684 (1903.5435 kb) | leading | -0.233 |
| 685 (1906.2415 kb) | 1267 (4170.51 kb) | NA | -0.18 | |
| AT-skew reverse | 1268 (0 kb) | 1898 (2709.9375 kb) | leading | 0.179 |
| 1899 (2714.6985 kb) | 2432 (4170.51 kb) | lagging | 0.243 |
More G than C on the leading strand for replication.