Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingopyxis.
Genome size (bp): 3345170
Number of genes 3165
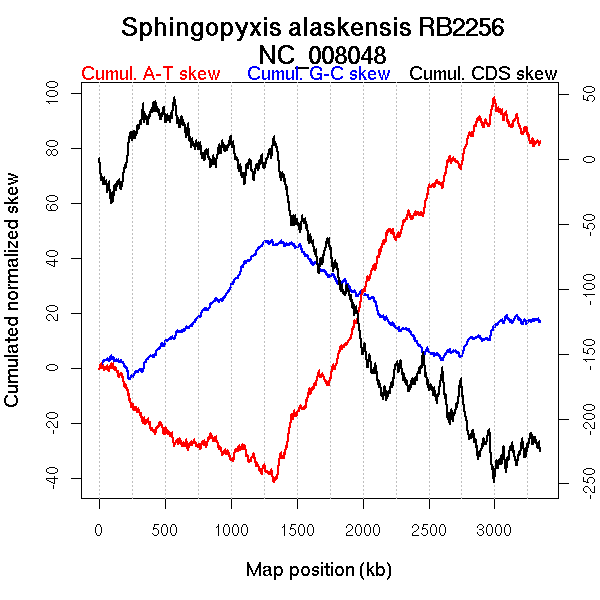
Oriloc predictions: Origin 2655 kb Terminus 1380 kb
Worning et al., 2006: Origin 2563 kb Terminus 1343 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 1.56 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.85 kb
Consensus predictions: Origin 2655 kb Terminus 1380 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 621 | 0.00667 | 1303 |
| GC-skew reverse | 2094 | 0.01 | 1331 |
| 2755 | 0.01333 | 2544 | |
| AT-skew forward | 806 | 0.03 | 1797 |
| 1278 | 0.00667 | 2952 | |
| AT-skew reverse | 2088 | 0.02 | 1302 |
| 2742 | 0.19333 | 2515 |
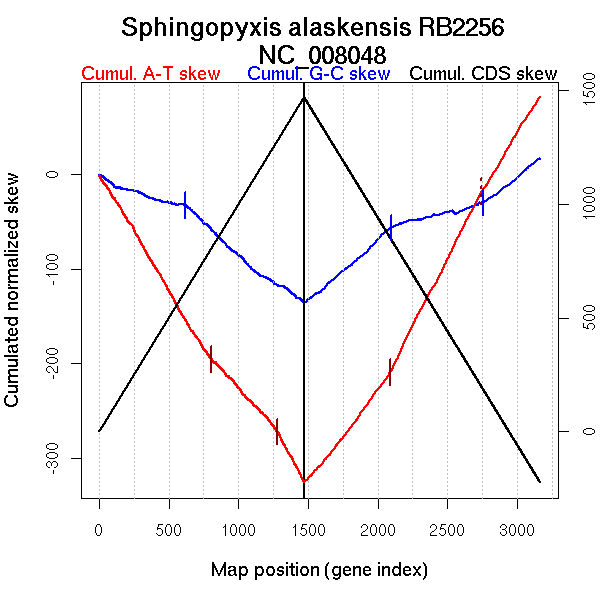
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 621 (1303.404 kb) | leading | -0.049 |
| 622 (1307.7945 kb) | 1471 (3344.839 kb) | NA | -0.121 | |
| GC-skew reverse | 1472 (0 kb) | 2094 (1330.5195 kb) | leading | 0.129 |
| 2095 (1331.943 kb) | 2755 (2544.049 kb) | lagging | 0.035 | |
| 2756 (2545.773 kb) | 3165 (3344.839 kb) | NA | 0.118 | |
| AT-skew forward | 1 (0 kb) | 806 (1797.4815 kb) | NA | -0.249 |
| 807 (1800.0655 kb) | 1278 (2951.6355 kb) | NA | -0.16 | |
| 1279 (2952.246 kb) | 1471 (3344.839 kb) | leading | -0.276 | |
| AT-skew reverse | 1472 (0 kb) | 2088 (1301.837 kb) | leading | 0.191 |
| 2089 (1303.6175 kb) | 2742 (2515.3615 kb) | lagging | 0.29 | |
| 2743 (2517.5575 kb) | 3165 (3344.839 kb) | NA | 0.246 |
More G than C on the leading strand for replication - for reverse encoded genes.