Rearranged Oriloc output example: Previous Index Next
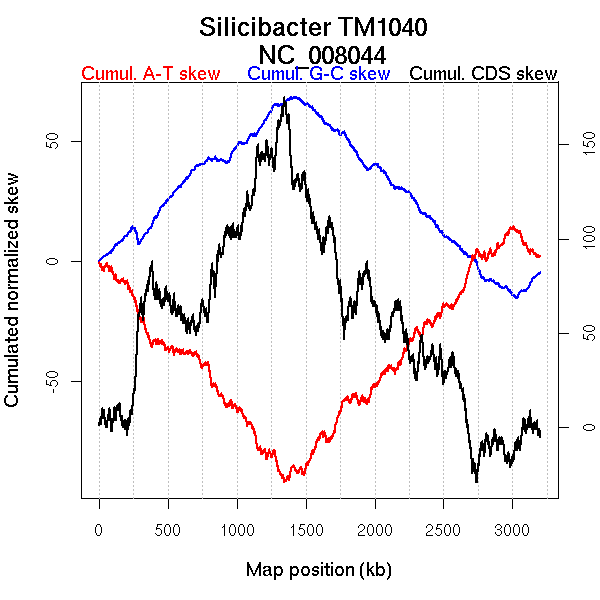
Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Silicibacter.
Genome size (bp): 3200938
Number of genes 3030
Oriloc predictions: Origin 3030 kb Terminus 1384 kb
Worning et al., 2006: Origin 3012 kb Terminus 1376 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 959.193 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.75 kb, 1957.88 kb
Consensus predictions: Origin 3030 kb Terminus 1384 kb
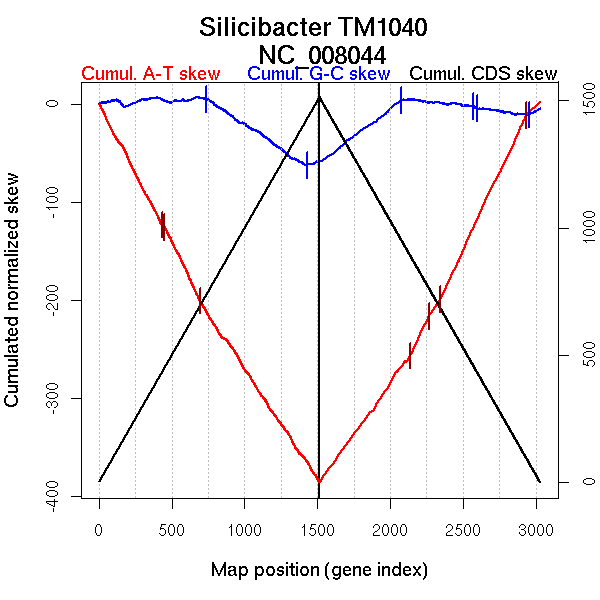
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 737 | 0 | 1433 |
| 1427 | 0 | 3034 | |
| GC-skew reverse | 2076 | 0.005 | 1382 |
| 2571 | 0.02667 | 2260 | |
| 2596 | 0.04 | 2302 | |
| 2952 | 0.01667 | 3039 | |
| AT-skew forward | 431 | 0 | 843 |
| 449 | 0 | 856 | |
| 692 | 0.03333 | 1313 | |
| AT-skew reverse | 2135 | 0.01167 | 1464 |
| 2265 | 0.02 | 1713 | |
| 2344 | 0.02167 | 1825 | |
| 2936 | 0.00167 | 2998 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 737 (1432.5035 kb) | leading | 0.006 |
| 738 (1433.265 kb) | 1427 (3034.0055 kb) | lagging | -0.1 | |
| 1428 (3035.889 kb) | 1514 (3200.382 kb) | leading | 0.05 | |
| GC-skew reverse | 1515 (0 kb) | 2076 (1382.016 kb) | leading | 0.113 |
| 2077 (1382.5055 kb) | 2571 (2259.748 kb) | lagging | -0.014 | |
| 2572 (2262.852 kb) | 2596 (2301.6385 kb) | lagging | -0.093 | |
| 2597 (2304.402 kb) | 2952 (3039.458 kb) | lagging | -0.02 | |
| 2953 (3040.3865 kb) | 3030 (3200.382 kb) | leading | 0.073 | |
| AT-skew forward | 1 (0 kb) | 431 (843.241 kb) | leading | -0.284 |
| 432 (843.762 kb) | 449 (855.898 kb) | leading | -0.121 | |
| 450 (856.4325 kb) | 692 (1313.266 kb) | leading | -0.31 | |
| 693 (1316.344 kb) | 1514 (3200.382 kb) | lagging | -0.223 | |
| AT-skew reverse | 1515 (0 kb) | 2135 (1463.669 kb) | leading | 0.208 |
| 2136 (1464.8365 kb) | 2265 (1713.4965 kb) | lagging | 0.318 | |
| 2266 (1714.12 kb) | 2344 (1825.3865 kb) | lagging | 0.214 | |
| 2345 (1826.4625 kb) | 2936 (2997.5475 kb) | lagging | 0.316 | |
| 2937 (3000.141 kb) | 3030 (3200.382 kb) | leading | 0.132 |
More G than C on the leading strand for replication.