Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Phyllobacteriaceae; Mesorhizobium.
Genome size (bp): 4412446
Number of genes 4064
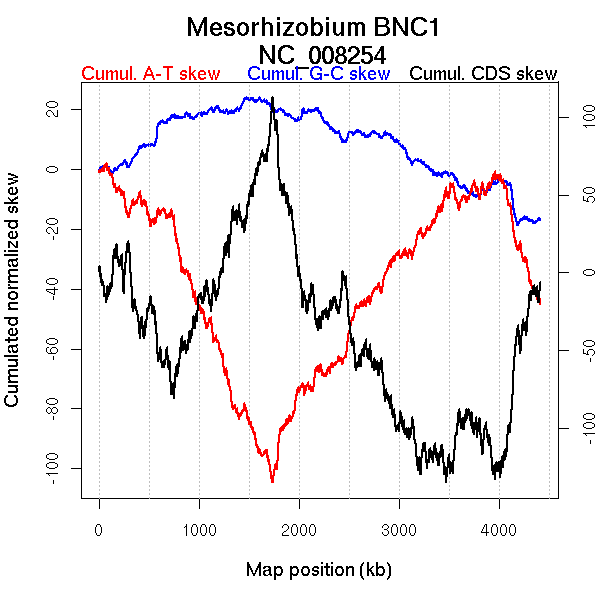
Oriloc predictions: Origin 3754 kb Terminus 1735 kb
Worning et al., 2006: Origin NA kb Terminus NA kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 3755.327 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.8 kb, 967.02 kb
Consensus predictions: Origin 3754 kb Terminus 1735 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 745 | 0 | 1540 |
| GC-skew reverse | 2750 | 0 | 1646 |
| AT-skew forward | 690 | 0 | 1456 |
| AT-skew reverse | 2778 | 0.01 | 1742 |
| 3938 | 0.01 | 4035 |
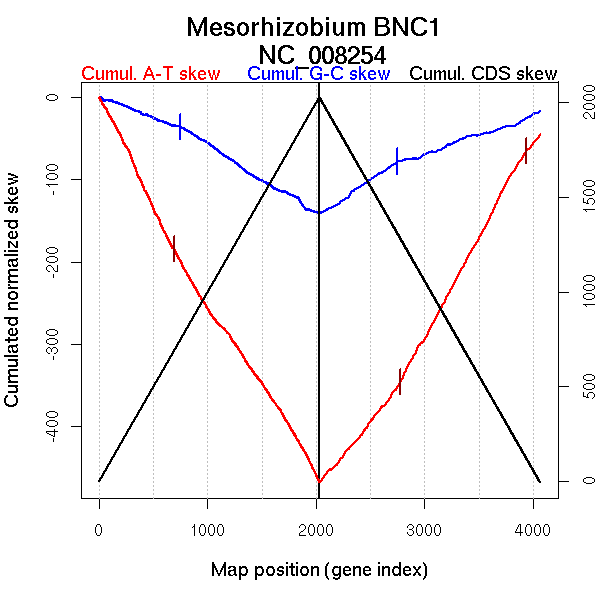
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 745 (1540.292 kb) | leading | -0.051 |
| 746 (1540.983 kb) | 2029 (4411.662 kb) | NA | -0.084 | |
| GC-skew reverse | 2030 (0 kb) | 2750 (1645.506 kb) | leading | 0.091 |
| 2751 (1646.596 kb) | 4064 (4411.662 kb) | NA | 0.046 | |
| AT-skew forward | 1 (0 kb) | 690 (1456.279 kb) | leading | -0.279 |
| 691 (1456.612 kb) | 2029 (4411.662 kb) | NA | -0.199 | |
| AT-skew reverse | 2030 (0 kb) | 2778 (1741.6505 kb) | leading | 0.162 |
| 2779 (1743.1055 kb) | 3938 (4035.232 kb) | lagging | 0.245 | |
| 3939 (4036.1585 kb) | 4064 (4411.662 kb) | leading | 0.16 |