Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Epsilonproteobacteria; Campylobacterales; Campylobacteraceae; Campylobacter.
Genome size (bp): 1641481
Number of genes 1623
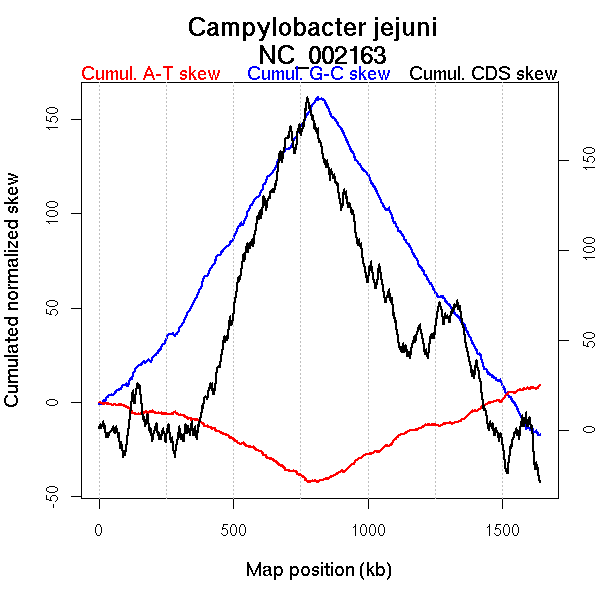
Oriloc predictions: Origin 0 kb Terminus 813 kb
Worning et al., 2006: Origin 1 kb Terminus 775 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 1262.127 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.66 kb
Consensus predictions: Origin 0 kb Terminus 813 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 486 | 0 | 830 |
| GC-skew reverse | 1120 | 0 | 818 |
| AT-skew forward | 495 | 0 | 861 |
| AT-skew reverse | 1126 | 0 | 829 |
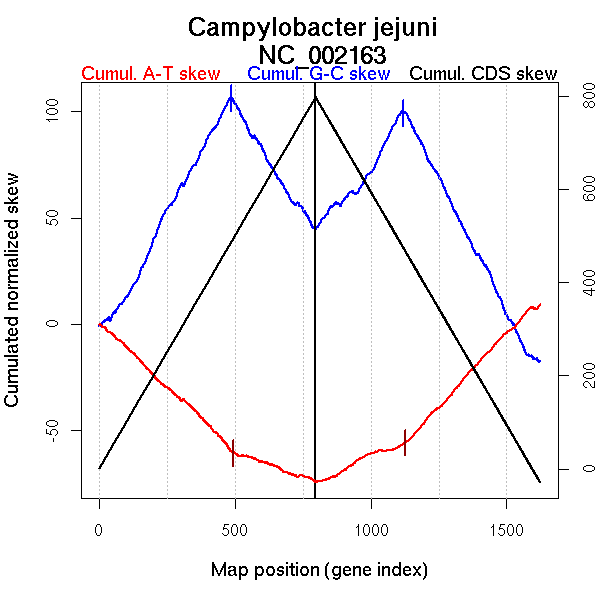
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 486 (830.069 kb) | leading | 0.237 |
| 487 (840.3345 kb) | 797 (1641.386 kb) | lagging | -0.2 | |
| GC-skew reverse | 798 (0 kb) | 1120 (818.14 kb) | leading | 0.165 |
| 1121 (818.866 kb) | 1623 (1641.386 kb) | lagging | -0.248 | |
| AT-skew forward | 1 (0 kb) | 495 (860.7225 kb) | leading | -0.121 |
| 496 (861.053 kb) | 797 (1641.386 kb) | lagging | -0.045 | |
| AT-skew reverse | 798 (0 kb) | 1126 (829.177 kb) | leading | 0.06 |
| 1127 (830.6035 kb) | 1623 (1641.386 kb) | lagging | 0.136 |
More G than C on the leading strand for replication.