Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Gammaproteobacteria; Pasteurellales; Pasteurellaceae; Pasteurella.
Genome size (bp): 2257487
Number of genes 2015
Oriloc predictions: Origin NA Terminus NA
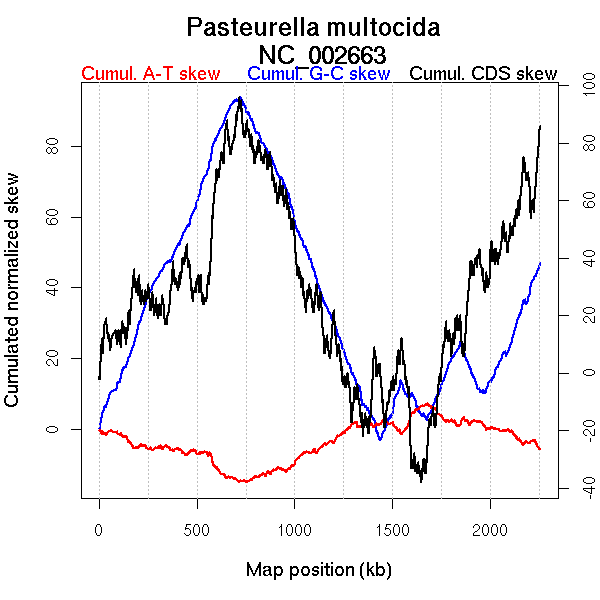
Worning et al., 2006: Origin 1671 kb Terminus 715 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 933.707 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 1357.68 kb
Consensus predictions: Origin 0 kb Terminus 715 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 352 | 0 | 714 |
| 610 | 0.00333 | 1408 | |
| 832 | 0.005 | 1864 | |
| 889 | 0 | 1967 | |
| GC-skew reverse | 1310 | 0 | 717 |
| 1666 | 0.00667 | 1426 | |
| 1862 | 0.02167 | 1851 | |
| 1907 | 0 | 1968 | |
| AT-skew forward | 335 | 0.04667 | 684 |
| AT-skew reverse | 1885 | 0.00667 | 1913 |
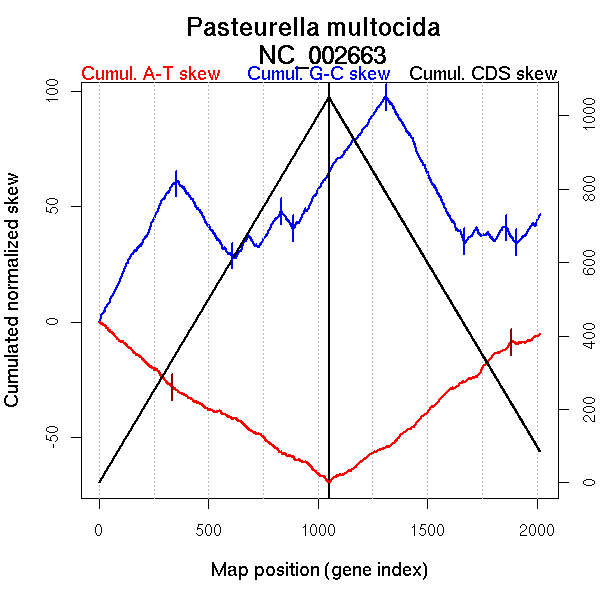
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 352 (713.9615 kb) | leading | 0.174 |
| 353 (714.737 kb) | 610 (1407.6845 kb) | lagging | -0.138 | |
| 611 (1408.2155 kb) | 832 (1863.9755 kb) | lagging | 0.075 | |
| 833 (1866.3625 kb) | 889 (1967.129 kb) | lagging | -0.136 | |
| 890 (1972.4515 kb) | 1050 (2257.487 kb) | lagging | 0.152 | |
| GC-skew reverse | 1051 (0 kb) | 1310 (717.3185 kb) | leading | 0.122 |
| 1311 (719.255 kb) | 1666 (1425.853 kb) | lagging | -0.182 | |
| 1667 (1428.7425 kb) | 1862 (1850.552 kb) | lagging | 0.014 | |
| 1863 (1851.7345 kb) | 1907 (1967.724 kb) | lagging | -0.149 | |
| 1908 (1969.5985 kb) | 2015 (2257.487 kb) | lagging | 0.11 | |
| AT-skew forward | 1 (0 kb) | 335 (683.782 kb) | leading | -0.083 |
| 336 (686.396 kb) | 1050 (2257.487 kb) | lagging | -0.056 | |
| AT-skew reverse | 1051 (0 kb) | 1885 (1912.779 kb) | NA | 0.074 |
| 1886 (1916.314 kb) | 2015 (2257.487 kb) | lagging | 0.029 |
More G than C on the leading strand for replication.