Rearranged Oriloc output example: Previous Index Next
Bacteria; Chlamydiae; Chlamydiales; Parachlamydiaceae; Candidatus Protochlamydia.
Genome size (bp): 2414465
Number of genes 2031
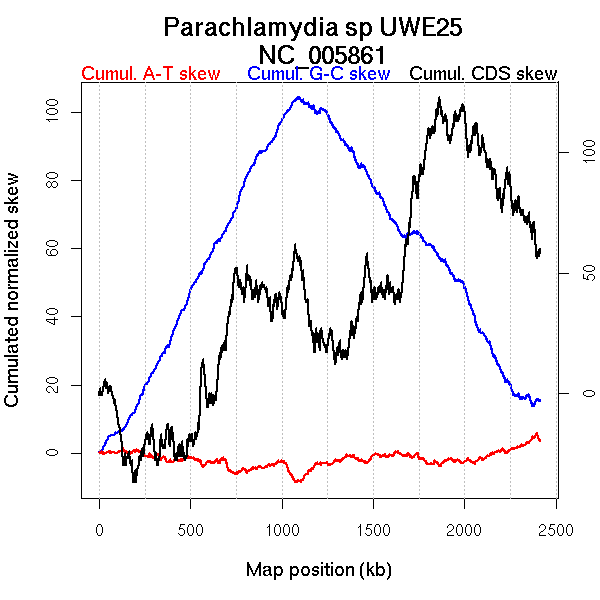
Oriloc predictions: Origin 0 kb Terminus 1092 kb
Worning et al., 2006: Origin 2401 kb Terminus 1092 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 636.887 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 424.97 kb, 1296.12 kb
Consensus predictions: Origin 0 kb Terminus 1092 kb
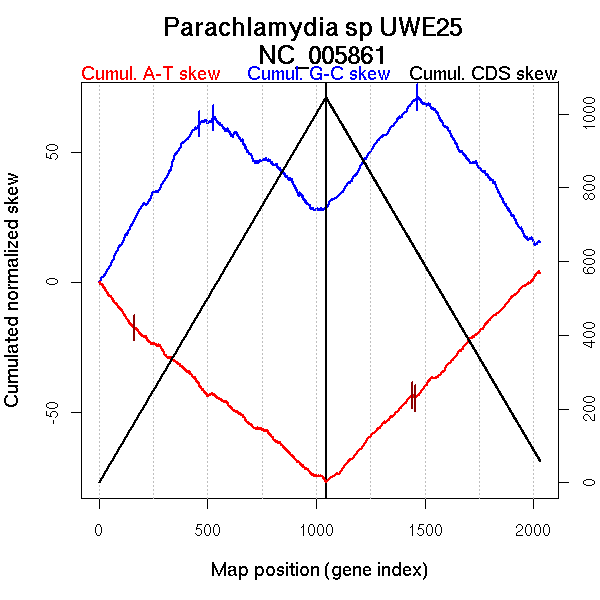
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 463 | 0.02 | 1058 |
| 527 | 0.03333 | 1216 | |
| GC-skew reverse | 1465 | 0 | 1087 |
| AT-skew forward | 160 | 0.00667 | 472 |
| AT-skew reverse | 1442 | 0.00667 | 1013 |
| 1454 | 0.00667 | 1050 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 463 (1058.05 kb) | leading | 0.133 |
| 464 (1060.165 kb) | 527 (1216.14 kb) | NA | 0.017 | |
| 528 (1221.702 kb) | 1045 (2414.389 kb) | lagging | -0.072 | |
| GC-skew reverse | 1046 (0 kb) | 1465 (1086.8535 kb) | leading | 0.104 |
| 1466 (1089.291 kb) | 2031 (2414.389 kb) | lagging | -0.107 | |
| AT-skew forward | 1 (0 kb) | 160 (472.4325 kb) | leading | -0.111 |
| 161 (475.734 kb) | 1045 (2414.389 kb) | NA | -0.067 | |
| AT-skew reverse | 1046 (0 kb) | 1442 (1012.8655 kb) | leading | 0.086 |
| 1443 (1015.5 kb) | 1454 (1049.5725 kb) | leading | -0.103 | |
| 1455 (1051.295 kb) | 2031 (2414.389 kb) | lagging | 0.085 |
More G than C on the leading strand for replication.