Rearranged Oriloc output example: Previous Index Next
Bacteria; Actinobacteria; Actinobacteridae; Actinomycetales; Corynebacterineae; Mycobacteriaceae; Mycobacterium; Mycobacterium avium complex (MAC).
Genome size (bp): 4829781
Number of genes 4350
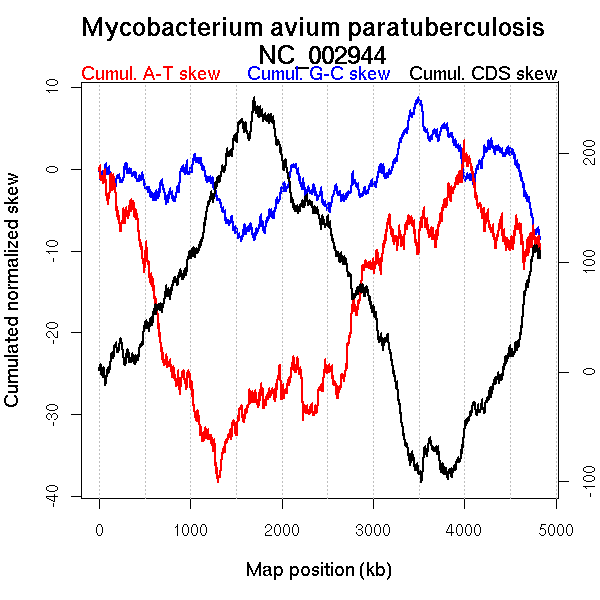
Oriloc predictions: Origin 3996 kb Terminus 1331 kb
Worning et al., 2006: Origin 3775 kb Terminus 1902 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 14.701 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 0.77 kb
Consensus predictions: Origin 0 kb Terminus 2500 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 658 | NA | 1216 |
| GC-skew reverse | 3037 | NA | 1997 |
| 4079 | NA | 4072 | |
| 4170 | NA | 4340 | |
| AT-skew forward | 609 | NA | 1145 |
| 1437 | NA | 3183 | |
| AT-skew reverse | 3370 | NA | 2685 |
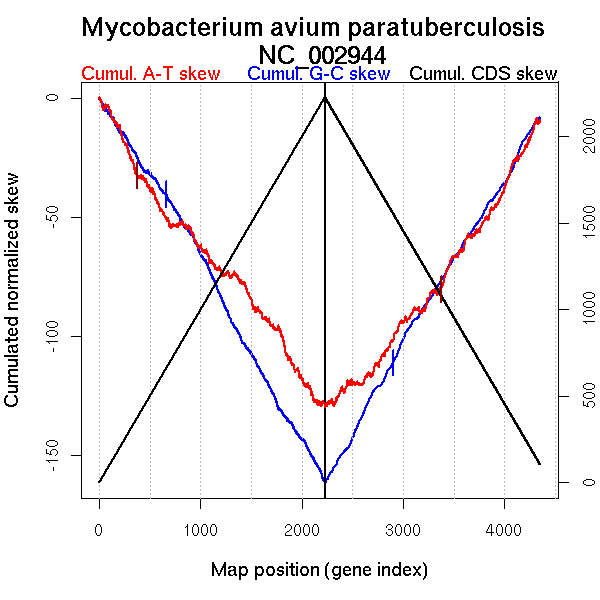
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 658 (1215.5775 kb) | leading | -0.063 |
| 659 (1218.5265 kb) | 2227 (4829.176 kb) | NA | -0.078 | |
| GC-skew reverse | 2228 (0 kb) | 3037 (1997.217 kb) | leading | 0.078 |
| 3038 (1999.698 kb) | 4079 (4072.397 kb) | NA | 0.066 | |
| 4080 (4074.437 kb) | 4170 (4340.153 kb) | lagging | 0.122 | |
| 4171 (4341.122 kb) | 4350 (4829.176 kb) | lagging | 0.063 | |
| AT-skew forward | 1 (0 kb) | 609 (1144.587 kb) | leading | -0.08 |
| 610 (1145.8255 kb) | 1437 (3183.0765 kb) | NA | -0.04 | |
| 1438 (3186.626 kb) | 2227 (4829.176 kb) | lagging | -0.067 | |
| AT-skew reverse | 2228 (0 kb) | 3370 (2685.1975 kb) | leading | 0.047 |
| 3371 (2686.451 kb) | 4350 (4829.176 kb) | lagging | 0.069 |