Rearranged Oriloc output example: Previous Index Next
Bacteria; Firmicutes; Clostridia; Clostridiales; Clostridiaceae; Clostridium.
Genome size (bp): 3031430
Number of genes 2660
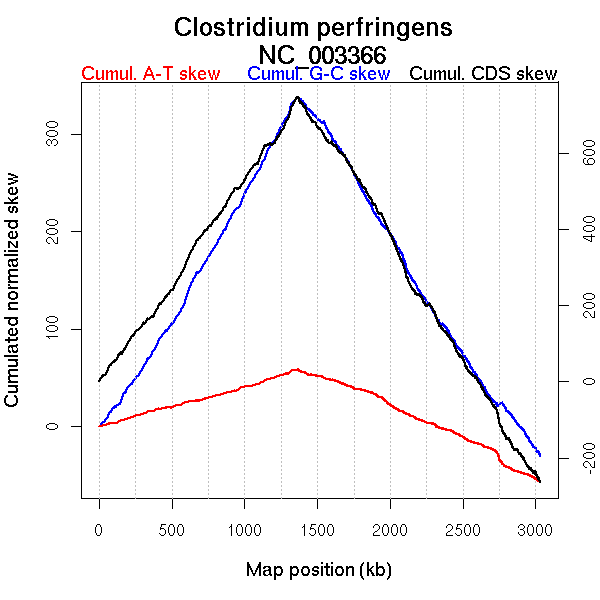
Oriloc predictions: Origin 0 kb Terminus 1359 kb
Worning et al., 2006: Origin 0 kb Terminus 1361 kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 0.143 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 1.1 kb, 2988.68 kb
Consensus predictions: Origin 0 kb Terminus 1359 kb
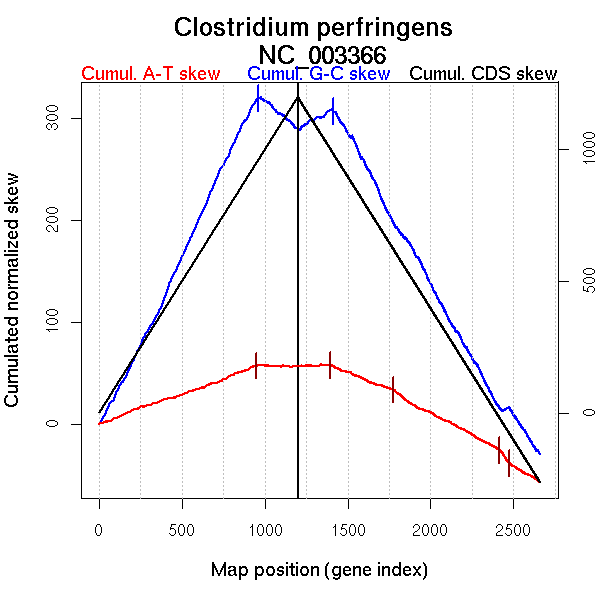
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 960 | NA | 1386 |
| GC-skew reverse | 1411 | NA | 1369 |
| AT-skew forward | 950 | NA | 1357 |
| AT-skew reverse | 1391 | NA | 1230 |
| 1771 | NA | 1885 | |
| 2416 | NA | 2728 | |
| 2475 | NA | 2773 |
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 960 (1385.8015 kb) | leading | 0.346 |
| 961 (1387.462 kb) | 1198 (3030.809 kb) | lagging | -0.135 | |
| GC-skew reverse | 1199 (0 kb) | 1411 (1369.3125 kb) | leading | 0.09 |
| 1412 (1369.9865 kb) | 2660 (3030.809 kb) | lagging | -0.279 | |
| AT-skew forward | 1 (0 kb) | 950 (1356.7575 kb) | leading | 0.059 |
| 951 (1357.074 kb) | 1198 (3030.809 kb) | lagging | -0.005 | |
| AT-skew reverse | 1199 (0 kb) | 1391 (1230.337 kb) | leading | 0.005 |
| 1392 (1233.2145 kb) | 1771 (1884.558 kb) | lagging | -0.064 | |
| 1772 (1885.0355 kb) | 2416 (2728.434 kb) | lagging | -0.087 | |
| 2417 (2730.371 kb) | 2475 (2773.477 kb) | lagging | -0.233 | |
| 2476 (2774.6675 kb) | 2660 (3030.809 kb) | lagging | -0.093 |
More G than C on the leading strand for replication.