Rearranged Oriloc output example: Previous Index Next
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae; Yersinia.
Genome size (bp): 4534590
Number of genes 3981
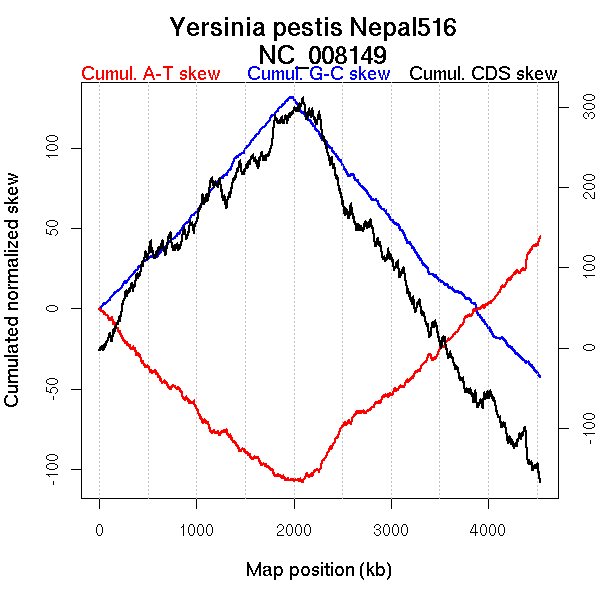
Oriloc predictions: Origin 0 kb Terminus 1981 kb
Worning et al., 2006: Origin NA kb Terminus NA kb
Position of the first peak in DnaA box distribution (Mackiewicz et al., 2004): 4432.612 kb
Position(s) of the dnaA gene(s) (Mackiewicz et al., 2004): 4499.9 kb
Consensus predictions: Origin 0 kb Terminus 1981 kb
Significant breakpoints (p-value <0.05):
| Breakpoints | P-value | Chromosome coordinate (kb) | |
| GC-skew forward | 1023 | NA | 1985 |
| GC-skew reverse | 2628 | NA | 1956 |
| AT-skew forward | 554 | NA | 1077 |
| 872 | NA | 1715 | |
| 1430 | NA | 3123 | |
| AT-skew reverse | 2663 | NA | 2047 |
| 3634 | NA | 3876 |
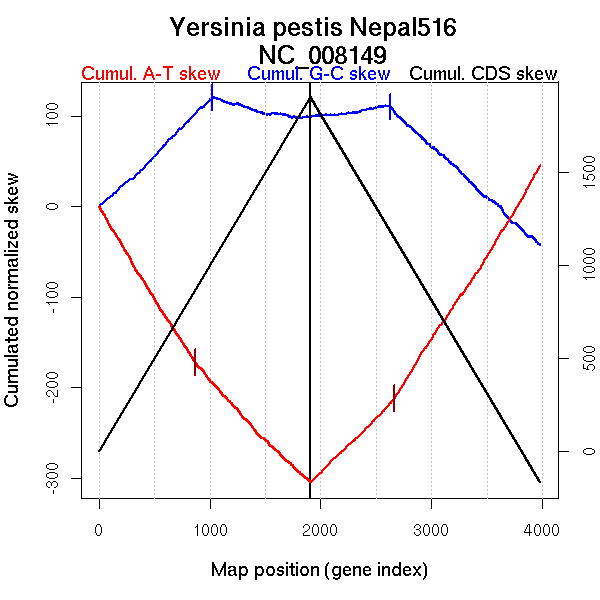
| From (index/coord) | To (index/coord) | Strand | Slope | |
| GC-skew forward | 1 (0 kb) | 1023 (1984.8975 kb) | leading | 0.12 |
| 1024 (1988.202 kb) | 1907 (4534.479 kb) | lagging | -0.026 | |
| GC-skew reverse | 1908 (0 kb) | 2628 (1955.8275 kb) | leading | 0.018 |
| 2629 (1958.0275 kb) | 3981 (4534.479 kb) | lagging | -0.112 | |
| AT-skew forward | 1 (0 kb) | 554 (1076.537 kb) | leading | -0.204 |
| 555 (1082.422 kb) | 872 (1714.5725 kb) | leading | -0.187 | |
| 873 (1716.009 kb) | 1430 (3122.5275 kb) | NA | -0.137 | |
| 1431 (3125.8055 kb) | 1907 (4534.479 kb) | lagging | -0.114 | |
| AT-skew reverse | 1908 (0 kb) | 2663 (2046.5395 kb) | leading | 0.12 |
| 2664 (2047.682 kb) | 3634 (3875.7085 kb) | lagging | 0.186 | |
| 3635 (3876.5695 kb) | 3981 (4534.479 kb) | lagging | 0.214 |
More G than C on the leading strand for replication.